|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.80 |
12.45 |
-11.65 |
1 |
5 |
0 |
66 |
380.451 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
7.99 |
9.26 |
-10.55 |
1 |
5 |
0 |
70 |
380.451 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
7.99 |
9.35 |
-11 |
1 |
5 |
0 |
70 |
380.451 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.36 |
Binding ≤ 10μM
|
|
ACE2-1-E |
Angiotensin-converting Enzyme 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.04 |
7.83 |
-63.43 |
1 |
7 |
-1 |
87 |
374.392 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.04 |
7.05 |
-52.32 |
3 |
7 |
1 |
88 |
376.408 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.78 |
7.73 |
-77.5 |
3 |
7 |
1 |
94 |
376.408 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
1.76 |
-42.1 |
3 |
8 |
1 |
120 |
375.382 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.56 |
-1.9 |
-38.85 |
3 |
8 |
1 |
123 |
375.382 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.88 |
3.14 |
-23.66 |
2 |
8 |
0 |
115 |
374.374 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
13523524
-
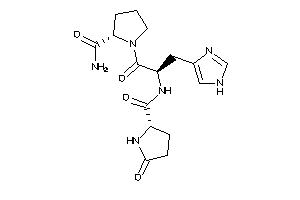
-
13523529
-
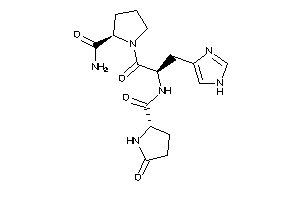
-
13523533
-
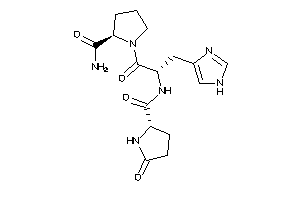
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.40 |
Binding ≤ 10μM
|
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 1μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 1μM
|
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 1μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 1μM
|
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
3 |
0.46 |
Functional ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
3 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.03 |
0.18 |
-23.59 |
5 |
10 |
0 |
150 |
362.39 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.03 |
0.01 |
-48.98 |
6 |
10 |
1 |
152 |
363.398 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.91 |
-8.71 |
-54.13 |
6 |
10 |
1 |
160 |
363.398 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
10 |
0.43 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.6 |
0.47 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
5.3 |
0.45 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
2.5 |
0.46 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
5.3 |
0.45 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
10 |
0.43 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.6 |
0.47 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
5.3 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
5.3 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
2.5 |
0.46 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.27 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1.92 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
1698429
-
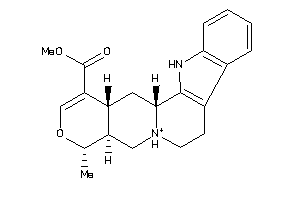
-
3978730
-
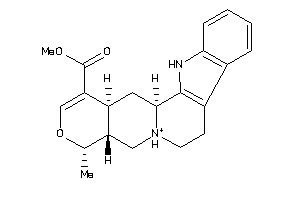
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.42 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.45 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
289 |
0.35 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80901-1-O |
HaCaT (Keratinocytes) (cluster #1 Of 2), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80901 |
Z80901
|
HaCaT (Keratinocytes) |
2100 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
1.59 |
5.79 |
-89.07 |
1 |
7 |
-2 |
127 |
342.303 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
4.25 |
-18.97 |
2 |
7 |
0 |
118 |
344.319 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
1.85 |
-45.9 |
1 |
6 |
-1 |
102 |
327.341 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
0.83 |
4.67 |
-49.37 |
1 |
6 |
-1 |
99 |
327.341 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
0.83 |
4.6 |
-53.01 |
1 |
6 |
-1 |
99 |
327.341 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
53 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
9.56 |
-39.99 |
1 |
4 |
1 |
37 |
324.448 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.36 |
-11.18 |
0 |
4 |
0 |
36 |
323.44 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.33 |
10.01 |
-90.93 |
2 |
4 |
2 |
39 |
325.456 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7400 |
0.31 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
5600 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.8 |
0.58 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.8 |
0.58 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1584.89319 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.8 |
0.58 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
120 |
0.46 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.8 |
0.58 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
120 |
0.46 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1584.89319 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
Z81047-1-O |
Human T-cell Line (cluster #1 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
0.22 |
-59.92 |
3 |
5 |
0 |
85 |
272.37 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.82 |
-1.07 |
-41.57 |
2 |
5 |
-1 |
80 |
271.362 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
0.05 |
-49.18 |
4 |
5 |
1 |
83 |
273.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
Z81047-1-O |
Human T-cell Line (cluster #1 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
0.82 |
-0.91 |
-41.79 |
2 |
5 |
-1 |
80 |
271.362 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
-3.36 |
2.41 |
-66.45 |
3 |
8 |
-1 |
127 |
267.265 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
2.7 |
-33.7 |
1 |
4 |
-1 |
65 |
253.347 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.48 |
2.59 |
-33.3 |
1 |
4 |
-1 |
65 |
253.347 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
4.52 |
-5.67 |
2 |
4 |
0 |
58 |
254.355 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
2.00 |
5.05 |
-60.11 |
1 |
5 |
0 |
78 |
250.261 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.55 |
6.46 |
-21.24 |
1 |
5 |
0 |
74 |
250.261 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.55 |
6.95 |
-50.92 |
2 |
5 |
1 |
75 |
251.269 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
320 |
0.51 |
Binding ≤ 1μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
320 |
0.51 |
Binding ≤ 1μM
|
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
320 |
0.51 |
Binding ≤ 10μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
320 |
0.51 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
350 |
0.50 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
3630 |
0.42 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1500 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.68 |
-19.84 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.18 |
8.8 |
-19.26 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
8.02 |
-6.12 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
320 |
0.51 |
Binding ≤ 1μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
320 |
0.51 |
Binding ≤ 1μM
|
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
320 |
0.51 |
Binding ≤ 10μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
320 |
0.51 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
350 |
0.50 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
3630 |
0.42 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1500 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.62 |
-19.9 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
8.24 |
-6.23 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
10.68 |
-5.25 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
5.93 |
-7.01 |
7 |
5 |
0 |
100 |
206.273 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.17 |
4.94 |
-28.01 |
7 |
5 |
1 |
104 |
206.273 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
5.35 |
-6.17 |
6 |
5 |
0 |
98 |
205.265 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|