|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
7.16 |
-37.11 |
1 |
5 |
-1 |
94 |
361.458 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.56 |
7.74 |
-38.77 |
0 |
5 |
-1 |
91 |
361.458 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
7.16 |
-11.18 |
1 |
5 |
0 |
89 |
362.466 |
7 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
50 |
1.46 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
50 |
1.46 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
50 |
1.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Lo
Low (pH 4.5-6)
|
0.75 |
3.78 |
-27.62 |
1 |
1 |
1 |
14 |
94.137 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1900 |
1.33 |
Binding ≤ 10μM |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5813 |
1.22 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
1900 |
1.33 |
Binding ≤ 10μM
|
|
TRPA1_RAT |
Q6RI86
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Rat |
5813 |
1.22 |
Functional ≤ 10μM |
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
1120 |
1.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
3.02 |
-27.34 |
1 |
1 |
1 |
14 |
100.166 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.25 |
2.45 |
-1.96 |
0 |
1 |
0 |
12 |
99.158 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.36 |
Binding ≤ 10μM
|
|
ACE2-1-E |
Angiotensin-converting Enzyme 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
-
21984495
-
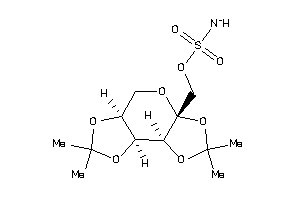
-
22061272
-
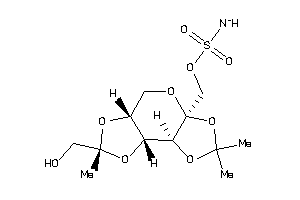
-
22061274
-
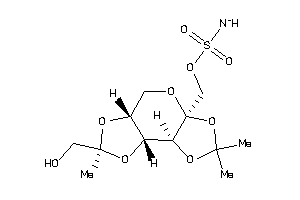
-
23586802
-
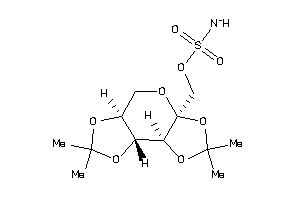
-
23586806
-
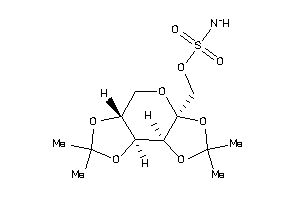
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-2-A |
Carbonic Anhydrase (cluster #2 Of 2), Archaea |
Archaea |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CYNT-3-B |
Carbonic Anhydrase (cluster #3 Of 3), Bacterial |
Bacteria |
474 |
0.40 |
Binding ≤ 10μM
|
|
P96878-2-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #2 Of 2), Bacterial |
Bacteria |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284-2-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #2 Of 2), Bacterial |
Bacteria |
612 |
0.40 |
Binding ≤ 10μM
|
|
B5SU02-4-E |
Alpha Carbonic Anhydrase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
C0IX24-3-E |
Carbonic Anhydrase (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
367 |
0.41 |
Binding ≤ 10μM
|
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-3-E |
Carbonic Anhydrase XIII (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14-5-E |
Carbonic Anhydrase XIV (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1460 |
0.37 |
Binding ≤ 10μM
|
|
CAH15-5-E |
Carbonic Anhydrase 15 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH3-4-E |
Carbonic Anhydrase III (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-7-E |
Carbonic Anhydrase IV (cluster #7 Of 16), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH5A-3-E |
Carbonic Anhydrase VA (cluster #3 Of 10), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5B-3-E |
Carbonic Anhydrase VB (cluster #3 Of 9), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-5-E |
Carbonic Anhydrase VII (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
58 |
0.46 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2-2-E |
Carbonic Anhydrase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4-2-E |
Carbonic Anhydrase IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Functional ≤ 10μM
|
|
CAN-2-F |
Carbonic Anhydrase (cluster #2 Of 3), Fungal |
Fungi |
110 |
0.44 |
Binding ≤ 10μM
|
|
Q3I4V7-4-F |
Carbonic Anhydrase 2 (cluster #4 Of 4), Fungal |
Fungi |
370 |
0.41 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
1110 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
29.1 |
0.48 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
10 |
0.51 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
474 |
0.40 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
110 |
0.44 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
367 |
0.41 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
120 |
0.44 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
172 |
0.43 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.45 |
Binding ≤ 1μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
370 |
0.41 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
15 |
0.50 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
58 |
0.46 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
63 |
0.46 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
25.4 |
0.48 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
25.4 |
0.48 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
63 |
0.46 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
45 |
0.47 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.87 |
0.58 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3.5 |
0.54 |
Binding ≤ 1μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
47 |
0.47 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
47 |
0.47 |
Binding ≤ 1μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
610 |
0.40 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
29.1 |
0.48 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
110 |
0.44 |
Binding ≤ 10μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
474 |
0.40 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
1020 |
0.38 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
367 |
0.41 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
10 |
0.51 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
172 |
0.43 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.45 |
Binding ≤ 10μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
370 |
0.41 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4900 |
0.34 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
1590 |
0.37 |
Binding ≤ 10μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
25.4 |
0.48 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
63 |
0.46 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
63 |
0.46 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
25.4 |
0.48 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.87 |
0.58 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3.5 |
0.54 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
1460 |
0.37 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
610 |
0.40 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
-1.52 |
-44.79 |
1 |
9 |
-1 |
113 |
338.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.16 |
-2 |
-12.26 |
2 |
9 |
0 |
116 |
339.366 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
7.65 |
-40.94 |
2 |
6 |
1 |
77 |
336.408 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
3.81 |
-7.79 |
1 |
6 |
0 |
76 |
335.4 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
4.66 |
-32 |
2 |
3 |
1 |
28 |
251.394 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
4.42 |
-28.28 |
2 |
3 |
1 |
28 |
251.394 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
6.27 |
-92.77 |
3 |
3 |
2 |
29 |
252.402 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Binding ≤ 10μM
|
|
ACM2-6-E |
Muscarinic Acetylcholine Receptor M2 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
9150 |
0.64 |
Binding ≤ 10μM |
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.67 |
Binding ≤ 10μM
|
|
ACM4-6-E |
Muscarinic Acetylcholine Receptor M4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.79 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM3-3-E |
Muscarinic Acetylcholine Receptor M3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
270 |
0.84 |
Binding ≤ 10μM
|
|
Z104303-5-O |
Muscarinic Acetylcholine Receptor (cluster #5 Of 7), Other |
Other |
77 |
0.91 |
Binding ≤ 10μM |
|
Z50512-4-O |
Cavia Porcellus (cluster #4 Of 7), Other |
Other |
600 |
0.79 |
Functional ≤ 10μM
|
|
Z50592-8-O |
Oryctolagus Cuniculus (cluster #8 Of 8), Other |
Other |
545 |
0.80 |
Functional ≤ 10μM |
|
Z80024-2-O |
A9 (Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1200 |
0.75 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
1.9 |
1.11 |
Binding ≤ 1μM
|
|
ACM1_MOUSE |
P12657
|
Muscarinic Acetylcholine Receptor M1, Mouse |
3.33 |
1.08 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
115 |
0.88 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
25 |
0.97 |
Binding ≤ 1μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
100 |
0.89 |
Binding ≤ 1μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
130 |
0.88 |
Binding ≤ 1μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
600 |
0.79 |
Binding ≤ 1μM
|
|
ACH10_RAT |
Q9JLB5
|
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
ACHA3_RAT |
P04757
|
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
ACHA4_RAT |
P09483
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
270 |
0.84 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
1.9 |
1.11 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM1_MOUSE |
P12657
|
Muscarinic Acetylcholine Receptor M1, Mouse |
3.33 |
1.08 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
115 |
0.88 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
1800 |
0.73 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
100 |
0.89 |
Binding ≤ 10μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
130 |
0.88 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
600 |
0.79 |
Binding ≤ 10μM
|
|
ACH10_RAT |
Q9JLB5
|
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3_RAT |
P04757
|
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
ACHA4_RAT |
P09483
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
270 |
0.84 |
Binding ≤ 10μM
|
|
Z80024 |
Z80024
|
A9 (Fibroblast Cells) |
1200 |
0.75 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
150 |
0.87 |
Functional ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
545 |
0.80 |
Functional ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
10000 |
0.64 |
Functional ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
140 |
0.87 |
Functional ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
73 |
0.91 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
545 |
0.80 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
3.10 |
6.38 |
-71.36 |
2 |
8 |
-1 |
132 |
402.433 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
16.27 |
-33.75 |
0 |
3 |
1 |
18 |
426.665 |
12 |
↓
|
|
|
|
Analogs
-
25072404
-
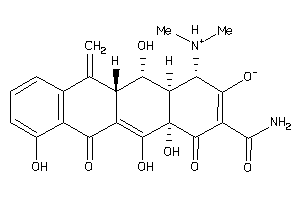
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MDFA-1-B |
Multidrug Translocase MdfA (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.75 |
-2.38 |
-66.56 |
6 |
10 |
-1 |
184 |
441.416 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.02 |
-1.44 |
-112.17 |
5 |
10 |
-2 |
187 |
440.408 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.75 |
-1.43 |
-155.4 |
5 |
10 |
-2 |
187 |
440.408 |
1 |
↓
|
|
|
|
Analogs
-
915591
-
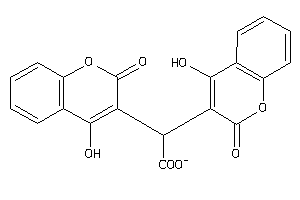
-
19336068
-
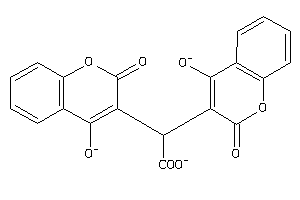
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
10.32 |
-144.54 |
0 |
8 |
-2 |
133 |
406.346 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.17 |
9.49 |
-67.48 |
1 |
8 |
-1 |
130 |
407.354 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.17 |
9.76 |
-70.02 |
1 |
8 |
-1 |
130 |
407.354 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
-1.04 |
-40.25 |
1 |
5 |
-1 |
82 |
231.231 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.80 |
1.52 |
-7.19 |
2 |
5 |
0 |
75 |
232.239 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
-0.99 |
-40.34 |
1 |
5 |
-1 |
82 |
231.231 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
-3.58 |
-42.94 |
0 |
5 |
-1 |
79 |
162.146 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.01 |
-4.34 |
-14.5 |
1 |
5 |
0 |
76 |
163.154 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.19 |
-2.09 |
-11.04 |
1 |
5 |
0 |
72 |
163.154 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-7-E |
Cyclooxygenase-2 (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
3.15 |
8.88 |
-8.32 |
1 |
5 |
0 |
94 |
361.458 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
11.75 |
-54.34 |
1 |
5 |
0 |
57 |
388.895 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.32 |
11.36 |
-60.96 |
1 |
5 |
0 |
57 |
388.895 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.32 |
9.49 |
-48.34 |
0 |
5 |
-1 |
56 |
387.887 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
11.77 |
-54.58 |
1 |
5 |
0 |
57 |
388.895 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.32 |
11.3 |
-61.05 |
1 |
5 |
0 |
57 |
388.895 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.32 |
9.46 |
-48.26 |
0 |
5 |
-1 |
56 |
387.887 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
12404993
-
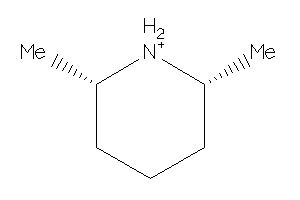
-
1683616
-
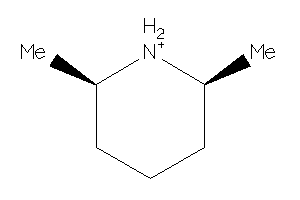
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
3.56 |
-31.69 |
2 |
1 |
1 |
17 |
114.212 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
-0.54 |
-42.79 |
1 |
4 |
-1 |
69 |
178.233 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
34566357
-
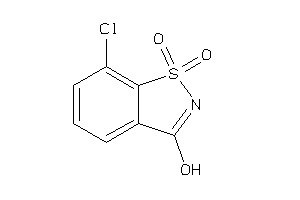
-
34588836
-
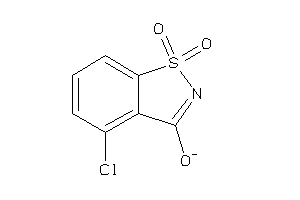
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
40.3 |
0.86 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
736 |
0.72 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
792 |
0.71 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
104 |
0.81 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
103 |
0.82 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
935 |
0.70 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
633 |
0.72 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
40.3 |
0.86 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
5950 |
0.61 |
Binding ≤ 10μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
792 |
0.71 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
104 |
0.81 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
5950 |
0.61 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
103 |
0.82 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
935 |
0.70 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
633 |
0.72 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
7960 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
-2.72 |
-47.81 |
0 |
4 |
-1 |
70 |
182.18 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.30 |
-3.48 |
-15.16 |
1 |
4 |
0 |
67 |
183.188 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
120 |
1.61 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
120 |
1.61 |
Binding ≤ 10μM
|
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
120 |
1.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.66 |
2.46 |
-34.4 |
2 |
1 |
1 |
17 |
86.158 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
388396
-
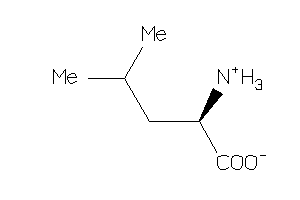
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.38 |
2.28 |
-37.79 |
3 |
3 |
0 |
68 |
131.175 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.80 |
-4.36 |
-57.03 |
3 |
4 |
0 |
85 |
125.149 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH4-6-E |
Carbonic Anhydrase IV (cluster #6 Of 16), Eukaryotic |
Eukaryotes |
99 |
0.75 |
Binding ≤ 10μM
|
|
AROQ-1-B |
3-dehydroquinate Dehydratase (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.98 |
3.22 |
-217.8 |
1 |
7 |
-3 |
141 |
189.099 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.98 |
1.24 |
-131.87 |
2 |
7 |
-2 |
138 |
190.107 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.98 |
-0.74 |
-64.58 |
3 |
7 |
-1 |
135 |
191.115 |
5 |
↓
|
|
|
|
Analogs
-
1148
-
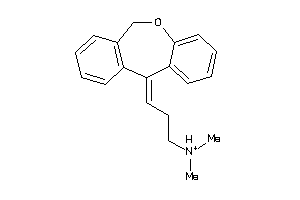
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
3162 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1258.92541 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
11.13 |
-37.52 |
1 |
2 |
1 |
14 |
280.391 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
56950
-
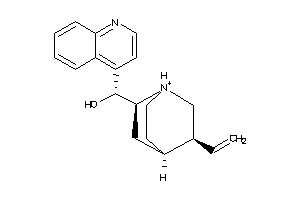
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.37 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
51 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
8.23 |
-43.02 |
2 |
3 |
1 |
38 |
295.406 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.03 |
8.66 |
-91.1 |
3 |
3 |
2 |
39 |
296.414 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
416
-
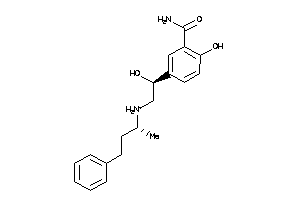
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
3.42 |
-60.25 |
6 |
5 |
1 |
100 |
329.42 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.85 |
4.43 |
-83.73 |
5 |
5 |
0 |
103 |
328.412 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
416
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
3.46 |
-59.33 |
6 |
5 |
1 |
100 |
329.42 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.85 |
4.47 |
-83.37 |
5 |
5 |
0 |
103 |
328.412 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
5283961
-
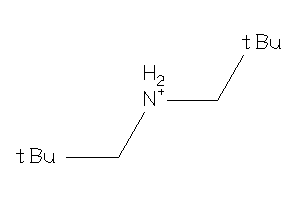
-
32219017
-
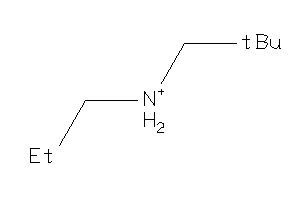
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
5.16 |
-33.98 |
2 |
1 |
1 |
17 |
130.255 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.36 |
-9.35 |
0 |
3 |
0 |
46 |
173.171 |
1 |
↓
|
|