|
|
Analogs
-
896455
-
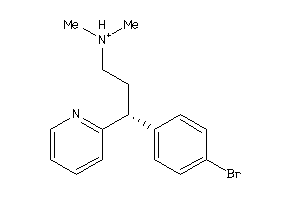
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.9 |
-40.9 |
1 |
2 |
1 |
17 |
320.254 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
2041666
-
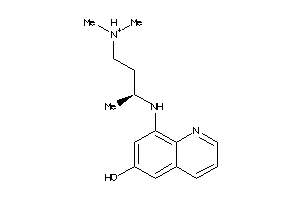
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
-1.64 |
-38.72 |
3 |
4 |
1 |
49 |
260.361 |
5 |
↓
|
|
|
|
Analogs
-
1576406
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
4.69 |
-10.97 |
3 |
6 |
0 |
92 |
336.775 |
7 |
↓
|
|
|
|
Analogs
-
4212654
-
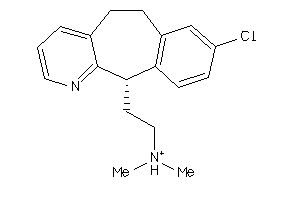
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
10.89 |
-33.55 |
1 |
2 |
1 |
17 |
301.841 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.80 |
11.17 |
-100.52 |
2 |
2 |
2 |
19 |
302.849 |
3 |
↓
|
|
|
|
Analogs
-
11616987
-
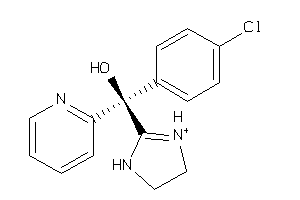
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
-4.85 |
-26.78 |
3 |
4 |
1 |
59 |
288.758 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
11616995
-
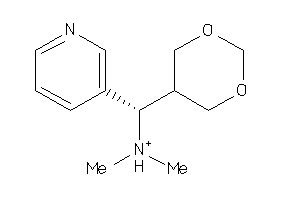
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
0.05 |
-40.42 |
1 |
4 |
1 |
35 |
223.296 |
3 |
↓
|
|
|
|
Analogs
-
7997952
-
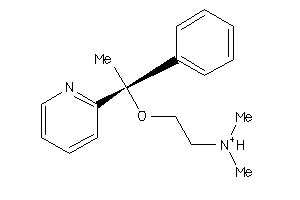
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
9.1 |
-35.1 |
1 |
3 |
1 |
27 |
271.384 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
1851415
-
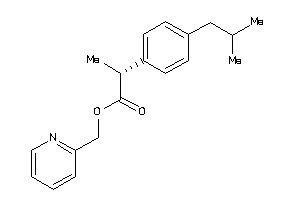
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
11.28 |
-8.36 |
0 |
3 |
0 |
39 |
297.398 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.86 |
3.04 |
-34.47 |
1 |
3 |
1 |
40 |
298.406 |
7 |
↓
|
|
|
|
Analogs
-
37868619
-
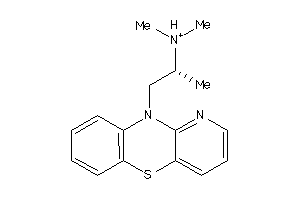
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
1.61 |
-39.61 |
1 |
3 |
1 |
22 |
286.424 |
3 |
↓
|
|
|
|
Analogs
-
1846399
-
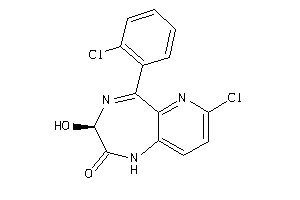
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
-5.55 |
-10.06 |
2 |
5 |
0 |
74 |
322.151 |
1 |
↓
|
|
|
|
Analogs
-
26736275
-
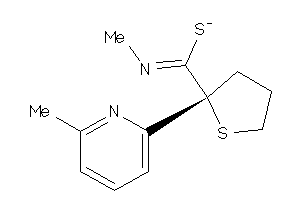
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
6.05 |
-42.97 |
0 |
2 |
-1 |
25 |
251.4 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
6.73 |
-14.46 |
1 |
2 |
0 |
27 |
252.408 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
6.21 |
-7.72 |
1 |
2 |
0 |
27 |
252.408 |
2 |
↓
|
|
|
|
Analogs
-
13282578
-
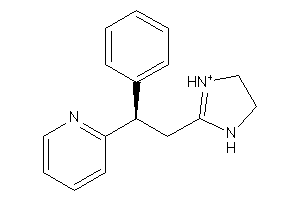
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
-2.26 |
-30.71 |
2 |
3 |
1 |
38 |
252.341 |
4 |
↓
|
|
|
|
Analogs
-
13209522
-
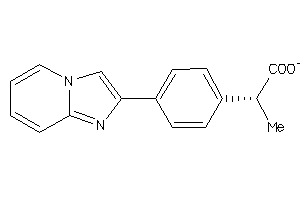
-
39220634
-
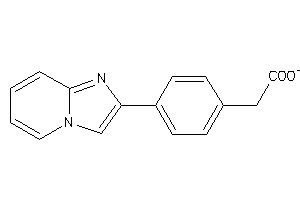
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
1.66 |
-55.07 |
0 |
4 |
-1 |
57 |
265.292 |
3 |
↓
|
|
|
|
Analogs
-
19702119
-
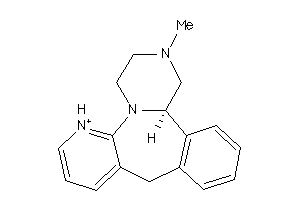
-
21981250
-
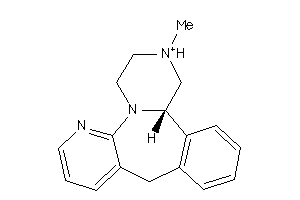
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
-0.77 |
-94.73 |
2 |
3 |
2 |
21 |
267.376 |
0 |
↓
|
|
|
|
Analogs
-
1843012
-
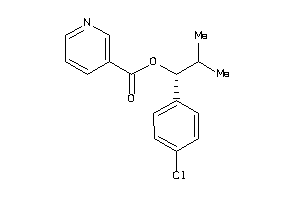
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
2.12 |
-5.74 |
0 |
3 |
0 |
39 |
289.762 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
1.56 |
-57.06 |
0 |
6 |
-1 |
84 |
323.328 |
4 |
↓
|
|
|
|
Analogs
-
4626674
-
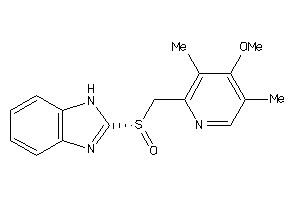
-
9084028
-
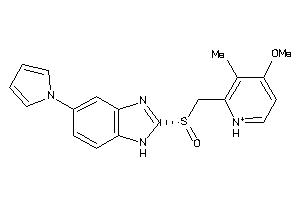
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
-3.53 |
-12.22 |
1 |
5 |
0 |
67 |
315.398 |
4 |
↓
|
|
|
|
Analogs
-
4626676
-
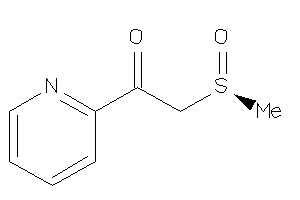
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.71 |
-1.67 |
-15.18 |
0 |
3 |
0 |
47 |
183.232 |
3 |
↓
|
|
|
|
Analogs
-
1684590
-
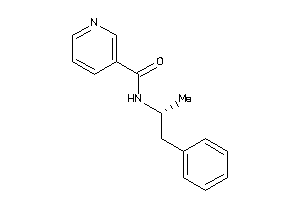
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
-0.15 |
-9.66 |
1 |
3 |
0 |
41 |
240.306 |
4 |
↓
|
|
|
|
Analogs
-
508068
-
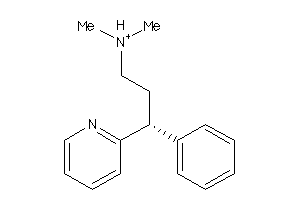
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
9.27 |
-38.74 |
1 |
2 |
1 |
17 |
241.358 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
26732916
-
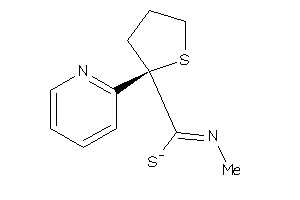
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
5.55 |
-42.41 |
0 |
2 |
-1 |
25 |
237.373 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.97 |
6.25 |
-13.49 |
1 |
2 |
0 |
27 |
238.381 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.97 |
5.64 |
-7.99 |
1 |
2 |
0 |
27 |
238.381 |
2 |
↓
|
|
|
|
Analogs
-
1846243
-
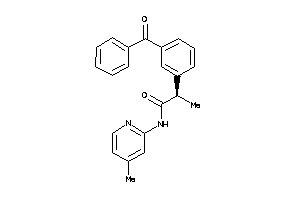
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.65 |
1.5 |
-16.85 |
1 |
4 |
0 |
59 |
344.414 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.65 |
1.15 |
-33.94 |
2 |
4 |
1 |
60 |
345.422 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
-1.7 |
-11.07 |
1 |
4 |
0 |
53 |
270.332 |
5 |
↓
|
|
|
|
Analogs
-
897261
-
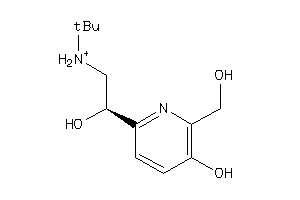
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
-3.63 |
-38.01 |
5 |
5 |
1 |
90 |
241.311 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.45 |
-2.87 |
-57.68 |
4 |
5 |
0 |
93 |
240.303 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.45 |
-2.81 |
-93.53 |
6 |
5 |
2 |
91 |
242.319 |
5 |
↓
|
|
|
|
Analogs
-
3959
-
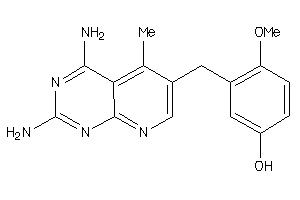
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
38 |
0.43 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.51 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.45 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
260 |
0.38 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 1μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 1μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 1μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 1μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 1μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 1μM
|
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 1μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 1μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 10μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 10μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 10μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 10μM
|
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 10μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 10μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 10μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815 |
Z80815
|
D54 Cell Line |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z80152 |
Z80152
|
HCT-8 (Ileocecal Adenocarcinoma) |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z81024 |
Z81024
|
NCI-H460 (Non-small Cell Lung Carcinoma) |
260 |
0.38 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z50339 |
Z50339
|
Pneumocystis Carinii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z80563 |
Z80563
|
U-373 MG ( Glioblastoma Cells) |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565 |
Z80565
|
U-87 MG (Glioblastoma Cells) |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583 |
Z80583
|
Vero (Kidney Cells) |
30 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.63 |
-16.02 |
4 |
7 |
0 |
109 |
325.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.41 |
-41.04 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.4 |
-28.87 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
|
|
Analogs
-
3831354
-
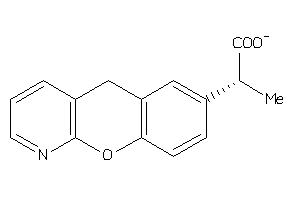
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.47 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
6.57 |
-50.99 |
0 |
4 |
-1 |
62 |
254.265 |
2 |
↓
|
|
|
|
Analogs
-
5141713
-
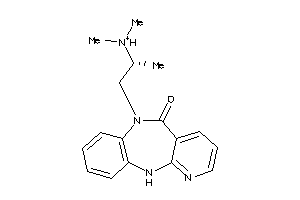
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
0.63 |
-29.22 |
2 |
5 |
1 |
55 |
297.382 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
2.6 |
-38.86 |
1 |
3 |
1 |
26 |
313.465 |
8 |
↓
|
|
|
|
Analogs
-
13232475
-
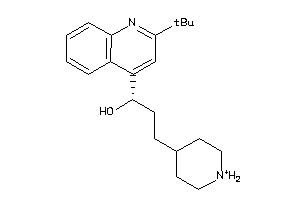
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
-2.42 |
-45.2 |
3 |
3 |
1 |
49 |
327.492 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
1582760
-
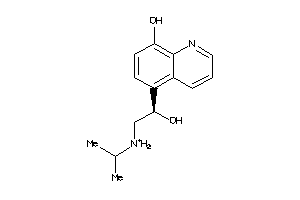
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
0.95 |
-50.11 |
4 |
4 |
1 |
70 |
247.318 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.58 |
-0.33 |
-9.25 |
3 |
4 |
0 |
65 |
246.31 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.58 |
1.71 |
-76.97 |
3 |
4 |
0 |
73 |
246.31 |
4 |
↓
|
|
|
|
Analogs
-
1842747
-
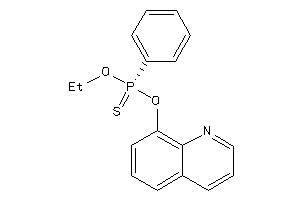
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
10.32 |
-18 |
0 |
3 |
0 |
31 |
329.361 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
2511.88643 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
8.33 |
-30.58 |
0 |
5 |
1 |
41 |
332.335 |
0 |
↓
|
|
|
|
Analogs
-
2041299
-
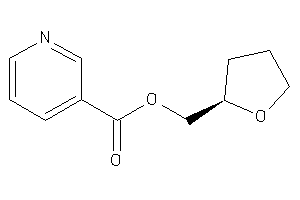
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
0.04 |
-7.73 |
0 |
4 |
0 |
48 |
207.229 |
4 |
↓
|
|
|
|
Analogs
-
4626699
-
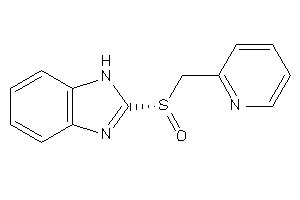
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.18 |
-4.5 |
-15.96 |
1 |
4 |
0 |
58 |
257.318 |
3 |
↓
|
|
|
|
Analogs
-
35899912
-
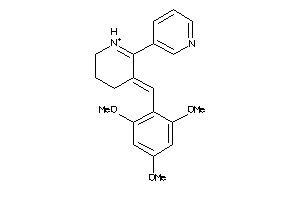
-
40950313
-
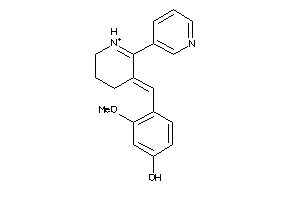
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
220 |
0.41 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
84 |
0.43 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
-1.23 |
-8.69 |
0 |
4 |
0 |
43 |
308.381 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
8.51 |
-14.6 |
2 |
6 |
0 |
74 |
400.482 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
8.99 |
-14.28 |
2 |
7 |
0 |
84 |
430.508 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
-2.5 |
-13.62 |
2 |
6 |
0 |
74 |
414.509 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.02 |
-2.39 |
-48.6 |
3 |
6 |
1 |
75 |
415.517 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
-3.63 |
-16.52 |
2 |
6 |
0 |
74 |
420.538 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.61 |
-3.52 |
-51.61 |
3 |
6 |
1 |
75 |
421.546 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
8.96 |
-22.06 |
2 |
7 |
0 |
84 |
430.508 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
9.4 |
-61.07 |
3 |
7 |
1 |
85 |
431.516 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
-2.91 |
-18.43 |
2 |
7 |
0 |
83 |
464.953 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.26 |
-2.8 |
-54.63 |
3 |
7 |
1 |
84 |
465.961 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
9.93 |
-17.41 |
2 |
6 |
0 |
74 |
428.536 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.23 |
10.37 |
-55.01 |
3 |
6 |
1 |
76 |
429.544 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
-2.08 |
-18.41 |
2 |
6 |
0 |
74 |
418.472 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.73 |
-1.97 |
-54.8 |
3 |
6 |
1 |
75 |
419.48 |
6 |
↓
|
|