|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
207 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
0.19 |
-9.4 |
1 |
3 |
0 |
46 |
300.398 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-1.85 |
-13.19 |
2 |
4 |
0 |
74 |
346.467 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
G6PD-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.36 |
Binding ≤ 10μM
|
|
Q9GRG7-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.76 |
-6.06 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1367 |
0.39 |
Binding ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
3380 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1367 |
0.39 |
Binding ≤ 10μM
|
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z50573 |
Z50573
|
Xenopus Laevis |
3380 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.9 |
-6.34 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
120294
-
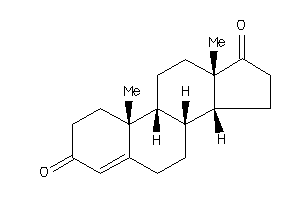
-
2046798
-
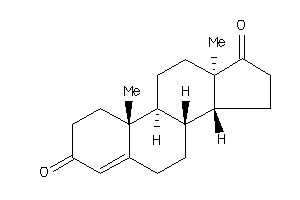
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.29 |
-10.68 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.06 |
9.39 |
-9.03 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
-3.47 |
-14.16 |
3 |
5 |
0 |
94 |
362.466 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.62 |
12.78 |
-1.53 |
1 |
1 |
0 |
20 |
386.664 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
490588
-
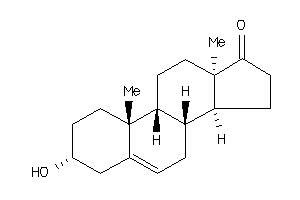
-
1036928
-
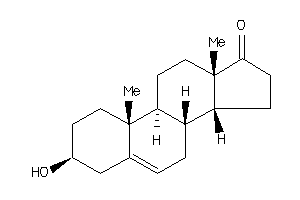
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.09 |
-6.2 |
1 |
2 |
0 |
37 |
288.431 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-9-E |
Acetylcholinesterase (cluster #9 Of 12), Eukaryotic |
Eukaryotes |
3 |
1.70 |
Binding ≤ 10μM
|
|
SC5A7-1-E |
High Affinity Choline Transporter 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1600 |
1.16 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.24 |
2.42 |
-27.92 |
1 |
2 |
1 |
20 |
104.173 |
2 |
↓
|
|
|
|
Analogs
-
1532640
-
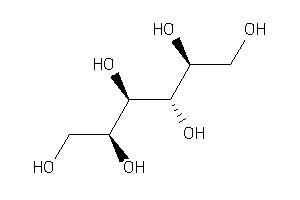
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.09 |
-12.44 |
-11.19 |
6 |
6 |
0 |
121 |
182.172 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
7.14 |
-7.63 |
1 |
2 |
0 |
37 |
270.372 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.53 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
|
SHBG-1-E |
Testis-specific Androgen-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Functional ≤ 10μM |
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Functional ≤ 10μM |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Functional ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z80226-1-O |
MCF7-2A (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 1μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 1μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 1μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 1μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 1μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 10μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 10μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.14 |
0.69 |
Functional ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
5.9 |
0.58 |
Functional ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
4.6 |
0.58 |
Functional ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
0.3 |
0.67 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
0.59 |
0.65 |
Functional ≤ 10μM
|
|
Z81068 |
Z81068
|
Ishikawa (Uterine Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80226 |
Z80226
|
MCF7-2A |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ADO_RAT |
Q9Z0U5
|
Aldehyde Oxidase, Rat |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
ADO_HUMAN |
Q06278
|
Aldehyde Oxidase, Human |
80 |
0.50 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
4.8 |
-5.19 |
2 |
2 |
0 |
40 |
272.388 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
11592545
-
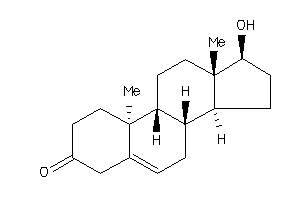
-
11592546
-
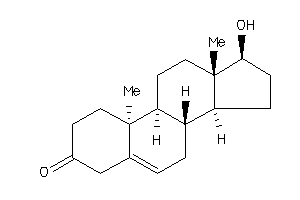
-
103270
-
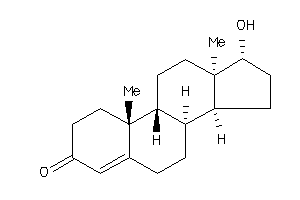
-
490793
-
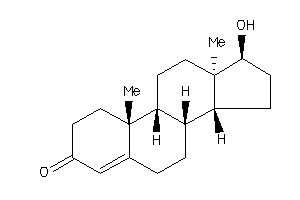
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 1μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 10μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 10μM
|
|
ERG2_YEAST |
P32352
|
C-8 Sterol Isomerase, Yeast |
7760 |
0.34 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
1200 |
0.39 |
Binding ≤ 10μM
|
|
S5A1_RAT |
P24008
|
Steroid 5-alpha-reductase 1, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
S5A2_RAT |
P31214
|
Steroid 5-alpha-reductase 2, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 10μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
3.2 |
0.57 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
10000 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.07 |
-7.05 |
1 |
2 |
0 |
37 |
288.431 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.99 |
5.16 |
-4.08 |
2 |
2 |
0 |
40 |
288.431 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.45 |
2.52 |
-31.62 |
3 |
5 |
1 |
76 |
265.362 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.45 |
2.98 |
-89.37 |
4 |
5 |
2 |
77 |
266.37 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.09 |
-11.21 |
-11.01 |
6 |
6 |
0 |
121 |
182.172 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
8637880
-
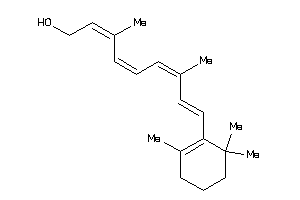
-
12484934
-
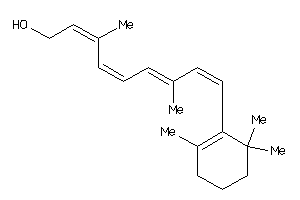
-
12496764
-
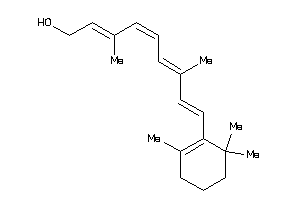
-
12496767
-
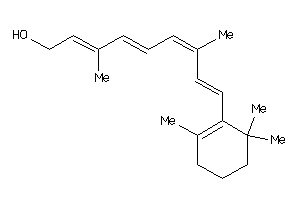
-
12496774
-
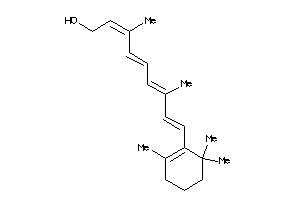
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB-1-E |
Beta-lactoglobulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1-1-E |
Cellular Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2-1-E |
Cellular Retinol-binding Protein II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3-1-E |
Interstitial Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4-1-E |
Plasma Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5-1-E |
Cellular Retinol-binding Protein III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7-1-E |
Cellular Retinol-binding Protein IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 1μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
9.9 |
-4.1 |
1 |
1 |
0 |
20 |
286.459 |
5 |
↓
|
|
|
|
Analogs
-
26301
-
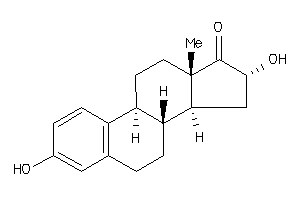
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
-2.91 |
-7.64 |
2 |
3 |
0 |
57 |
286.371 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
-2.73 |
-14.1 |
3 |
5 |
0 |
94 |
362.466 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
4.09 |
-5.74 |
2 |
3 |
0 |
58 |
304.43 |
0 |
↓
|
|
|
|
Analogs
-
6067733
-
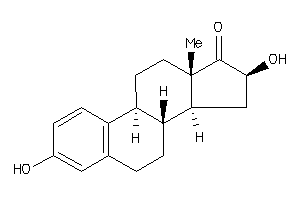
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
3.78 |
-7.98 |
2 |
3 |
0 |
58 |
286.371 |
0 |
↓
|
|
|
|
Analogs
-
4546563
-
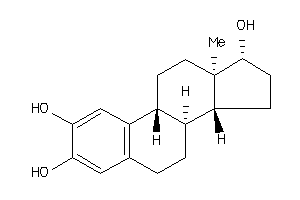
-
25757123
-
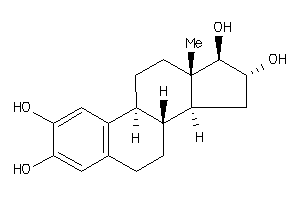
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
2.72 |
-7.57 |
3 |
3 |
0 |
61 |
288.387 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
-1.75 |
-10.23 |
2 |
3 |
0 |
57 |
286.371 |
0 |
↓
|
|
|
|
Analogs
-
585979
-
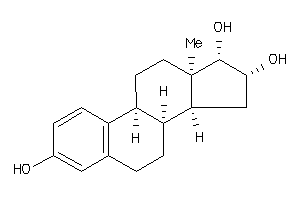
-
1736208
-
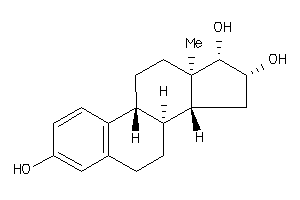
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
2.81 |
-7.02 |
3 |
3 |
0 |
61 |
288.387 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
585979
-

-
1736208
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
2.65 |
-6.17 |
3 |
3 |
0 |
61 |
288.387 |
0 |
↓
|
|
|
|
Analogs
-
3843834
-
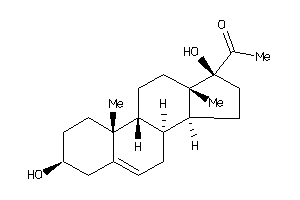
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.06 |
-5.89 |
2 |
3 |
0 |
58 |
332.484 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2620 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
2620 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.33 |
-5.82 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
4.6 |
-3.48 |
2 |
2 |
0 |
40 |
292.463 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
-0.14 |
-11.03 |
1 |
3 |
0 |
54 |
284.355 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
7.5 |
-9.66 |
1 |
3 |
0 |
54 |
330.468 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
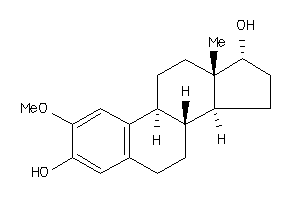
Analogs
-
3996018
-
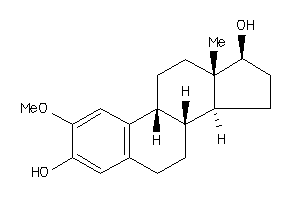
-
5855431
-
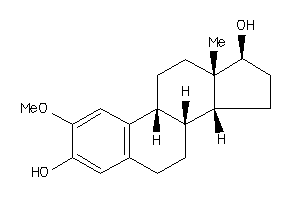
-
6482664
-
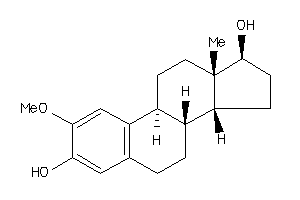
-
22062280
-
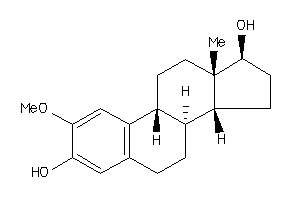
-
34019293
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.34 |
Binding ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.34 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.34 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.35 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.35 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
842 |
0.39 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
2650 |
0.35 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
867 |
0.39 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
840 |
0.39 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
500 |
0.40 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
1000 |
0.38 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
608 |
0.40 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
790 |
0.39 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 5), Other |
Other |
840 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
4.79 |
-6.89 |
2 |
3 |
0 |
50 |
302.414 |
1 |
↓
|
|
|
|
Analogs
-
4556638
-
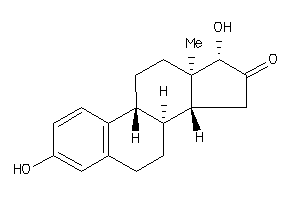
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
-2.73 |
-7.53 |
2 |
3 |
0 |
57 |
286.371 |
0 |
↓
|
|