|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
9 |
-14.93 |
0 |
4 |
0 |
39 |
277.364 |
8 |
↓
|
|
|
|
Analogs
-
1841718
-
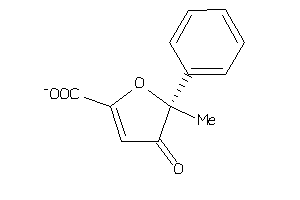
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HCAR2-1-E |
HM74 Nicotinic Acid GPCR (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM
|
|
HCAR3-1-E |
HM74 Nicotinic Acid GPCR (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.47 |
Binding ≤ 10μM
|
|
HCAR2-3-E |
HM74 Nicotinic Acid GPCR (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.55 |
Functional ≤ 10μM
|
|
HCAR3-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.57 |
-41.98 |
0 |
4 |
-1 |
66 |
217.2 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
-3.82 |
-11.37 |
4 |
7 |
0 |
99 |
303.366 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
-3.62 |
-38.89 |
5 |
7 |
1 |
100 |
304.374 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
-3.63 |
-39.25 |
5 |
7 |
1 |
100 |
304.374 |
5 |
↓
|
|
|
|
Analogs
-
4611316
-
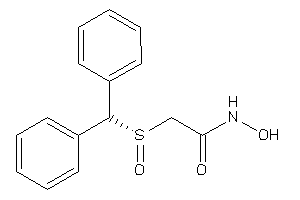
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.28 |
4.89 |
-24.55 |
2 |
4 |
0 |
66 |
289.356 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.28 |
5.63 |
-61.69 |
1 |
4 |
-1 |
69 |
288.348 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
4095934
-
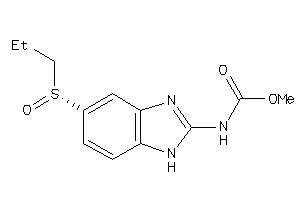
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
3.06 |
-23.33 |
2 |
6 |
0 |
87 |
281.337 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
1999253
-
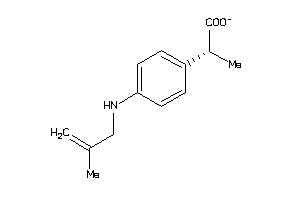
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
0.47 |
-48.57 |
1 |
3 |
-1 |
52 |
218.276 |
5 |
↓
|
|
|
|
Analogs
-
86599
-
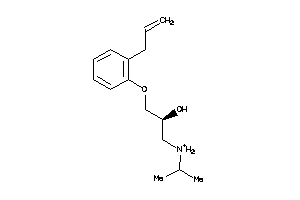
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1.51 |
0.69 |
Binding ≤ 1μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 1μM
|
|
ADRB2_BOVIN |
Q28044
|
Beta-2 Adrenergic Receptor, Bovin |
2.98 |
0.66 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1.51 |
0.69 |
Binding ≤ 1μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1.51 |
0.69 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1.51 |
0.69 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 10μM
|
|
ADRB2_BOVIN |
Q28044
|
Beta-2 Adrenergic Receptor, Bovin |
2.98 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1.51 |
0.69 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1.51 |
0.69 |
Binding ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
2.3 |
0.67 |
Binding ≤ 10μM
|
|
FDFT_RAT |
Q02769
|
Squalene Synthetase, Rat |
4200 |
0.42 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1995.26231 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.16 |
-40.25 |
3 |
3 |
1 |
46 |
250.362 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
3.88 |
-5.22 |
2 |
3 |
0 |
41 |
249.354 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
16090786
-
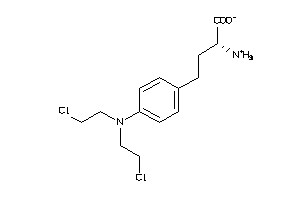
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
-1.91 |
-46.13 |
3 |
4 |
0 |
71 |
319.232 |
9 |
↓
|
|
|
|
Analogs
-
4211922
-
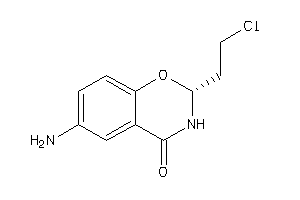
Draw
Identity
99%
90%
80%
70%
Popular Name:
2,3-Dihydro-6-amino-2-(2-chloroethyl)-4H-1,3-benzoxazin-4-one; 2-(beta-Chloroethyl)-2,3-dihydro-4-oxo-6-amino-1,3-benzoxazine; 2H-1,3-Benzoxazin-4(3H)-one, 6-amino-2-(2-chloroethyl)-; 4H-1,3-Benzoxazin-4-one, 2,3-dihydro-6-amino-2-(2-chloroethyl)-; 4H-1,3
2,3-Dihydro-6-amino-2-(2-chloroe…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
-4.5 |
-10.37 |
3 |
4 |
0 |
64 |
226.663 |
2 |
↓
|
|
|
|
Analogs
-
2020048
-
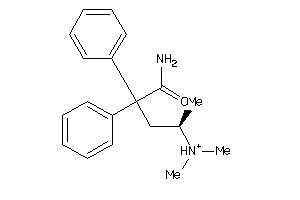
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
0.89 |
-37.97 |
3 |
3 |
1 |
47 |
297.422 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.89 |
2.55 |
-38.03 |
3 |
1 |
1 |
28 |
110.18 |
0 |
↓
|
|
|
|
Analogs
-
5211994
-
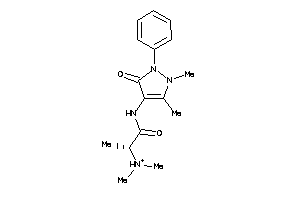
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
1.34 |
-47.14 |
2 |
6 |
1 |
60 |
303.386 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
1842933
-
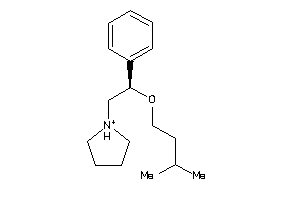
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
1.56 |
-33.37 |
1 |
2 |
1 |
13 |
262.417 |
7 |
↓
|
|
|
|
Analogs
-
164392
-
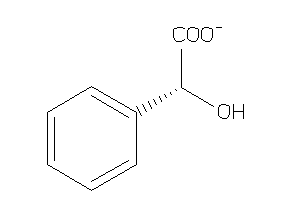
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
3.51 |
-45.3 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
-
2034893
-
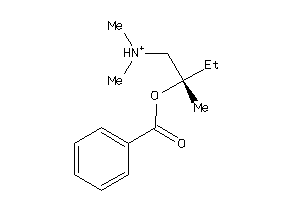
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
8.97 |
-38.55 |
1 |
3 |
1 |
31 |
236.335 |
6 |
↓
|
|
|
|
Analogs
-
5353020
-
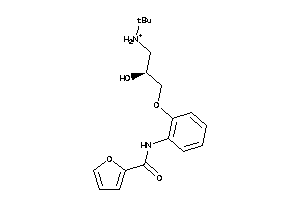
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
-2.45 |
-46.93 |
4 |
6 |
1 |
88 |
333.408 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
1844865
-
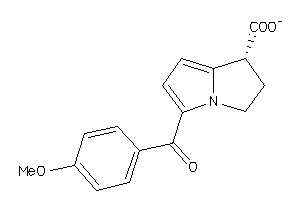
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
0.86 |
-47.56 |
0 |
5 |
-1 |
71 |
284.291 |
4 |
↓
|
|
|
|
Analogs
-
31828980
-
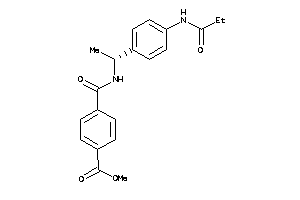
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
10.09 |
-18.5 |
2 |
6 |
0 |
85 |
402.45 |
7 |
↓
|
|
|
|
Analogs
-
1559439
-
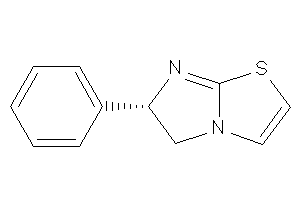
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
-1.57 |
-7.36 |
0 |
2 |
0 |
17 |
202.282 |
1 |
↓
|
|
|
|
Analogs
-
5138895
-
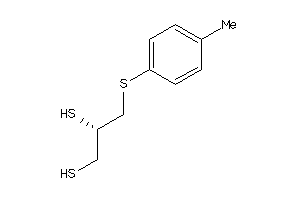
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
-0.45 |
-4.51 |
0 |
0 |
0 |
0 |
230.423 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
-1.41 |
-40.76 |
0 |
0 |
-1 |
0 |
229.415 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
-1.41 |
-41.83 |
0 |
0 |
-1 |
0 |
229.415 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
-0.2 |
-40.49 |
1 |
3 |
1 |
12 |
254.357 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
6.59 |
-56.84 |
0 |
4 |
-1 |
66 |
179.151 |
3 |
↓
|
|
|
|
Analogs
-
28101468
-
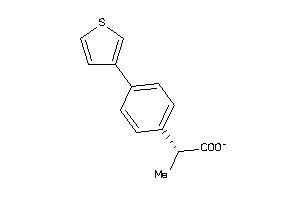
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
1.25 |
-48.65 |
0 |
2 |
-1 |
40 |
231.296 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
85733
-
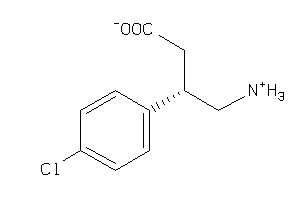
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.73 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.75 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.75 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB2-4-E |
GABA Receptor Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
Z104301-5-O |
GABA-A Receptor; Anion Channel (cluster #5 Of 8), Other |
Other |
30 |
0.75 |
Binding ≤ 10μM
|
|
Z50512-6-O |
Cavia Porcellus (cluster #6 Of 7), Other |
Other |
6400 |
0.52 |
Functional ≤ 10μM |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
330 |
0.65 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
1800 |
0.57 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
50 |
0.73 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
330 |
0.65 |
Binding ≤ 1μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
330 |
0.65 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
330 |
0.65 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
30 |
0.75 |
Binding ≤ 1μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
140 |
0.69 |
Binding ≤ 1μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
15 |
0.78 |
Binding ≤ 1μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
140 |
0.69 |
Binding ≤ 1μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
15 |
0.78 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
50 |
0.73 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
330 |
0.65 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
30 |
0.75 |
Binding ≤ 10μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
140 |
0.69 |
Binding ≤ 10μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
15 |
0.78 |
Binding ≤ 10μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
15 |
0.78 |
Binding ≤ 10μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
140 |
0.69 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
6400 |
0.52 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
330 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
4.81 |
-48.33 |
3 |
3 |
0 |
68 |
213.664 |
4 |
↓
|
|
|
|
Analogs
-
2007481
-
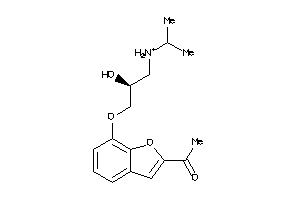
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
-1.86 |
-47.62 |
3 |
5 |
1 |
76 |
292.355 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
1999295
-
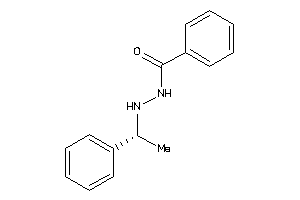
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
6.89 |
-9.89 |
2 |
3 |
0 |
41 |
240.306 |
4 |
↓
|
|
|
|
Analogs
-
1872015
-
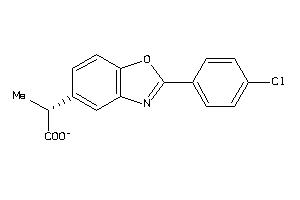
-
33420303
-
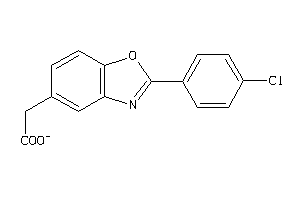
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
-0.61 |
-46.43 |
0 |
4 |
-1 |
66 |
300.721 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22AC-1-E |
Solute Carrier Family 22 Member 12 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22AC_HUMAN |
Q96S37
|
Solute Carrier Family 22 Member 12, Human |
2800 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
0.72 |
-9.64 |
1 |
3 |
0 |
50 |
266.296 |
3 |
↓
|
|
|
|
Analogs
-
5438128
-
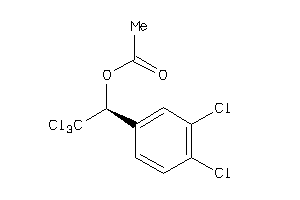
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
1.55 |
-4.97 |
0 |
2 |
0 |
26 |
336.429 |
4 |
↓
|
|
|
|
Analogs
-
6096699
-
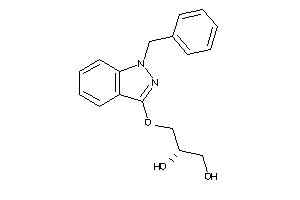
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
-5.31 |
-12.11 |
2 |
5 |
0 |
67 |
298.342 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
396186
-
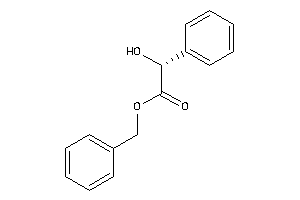
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
7.58 |
-6.66 |
1 |
3 |
0 |
47 |
242.274 |
5 |
↓
|
|
|
|
Analogs
-
338236
-
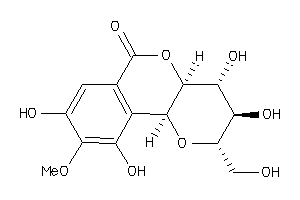
-
389665
-
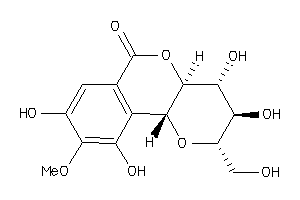
-
1638183
-
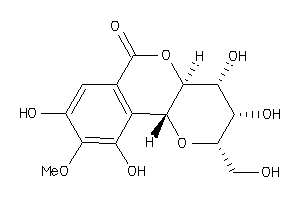
-
2048952
-
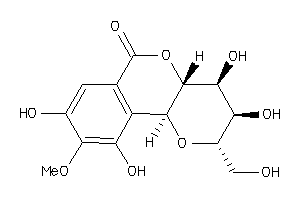
-
3871709
-
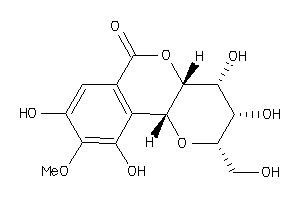
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3R,4S,4aR,10bR)-3,4,8,10-tetrahydroxy-9-methoxy-2-methylol-3,4,4a,10b-tetrahydro-2H-pyrano[3,2-c
(2S,3R,4S,4aR,10bR)-3,4,8,10-tet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.90 |
-7.1 |
-12.49 |
5 |
9 |
0 |
146 |
328.273 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
5764638
-
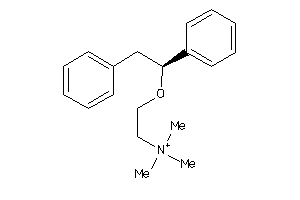
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
4.01 |
-31.08 |
0 |
2 |
1 |
9 |
284.423 |
7 |
↓
|
|
|
|
Analogs
-
6037202
-
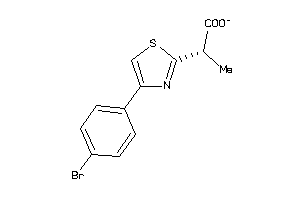
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-0.81 |
-45.8 |
0 |
3 |
-1 |
53 |
311.18 |
3 |
↓
|
|
|
|
Analogs
-
402865
-
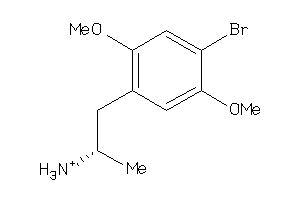
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.77 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.77 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3340 |
0.51 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.50 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3340 |
0.51 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.50 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3340 |
0.51 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.50 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.69 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.71 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
63 |
0.67 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.68 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.67 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.72 |
Functional ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.67 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
6.1 |
0.77 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
145 |
0.64 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
1.9 |
0.81 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
145 |
0.64 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
145 |
0.64 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
5000 |
0.49 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
5000 |
0.49 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
5000 |
0.49 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
5000 |
0.49 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
1.9 |
0.81 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
145 |
0.64 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
145 |
0.64 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
145 |
0.64 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
22 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
3.72 |
-40.05 |
3 |
3 |
1 |
46 |
275.166 |
4 |
↓
|
|