|
|
Analogs
-
538404
-
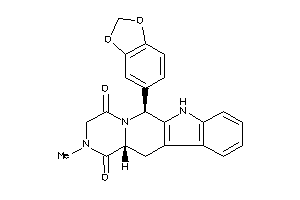
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
6.9 |
-12.89 |
1 |
7 |
0 |
75 |
389.411 |
1 |
↓
|
|
|
|
Analogs
-
8204637
-
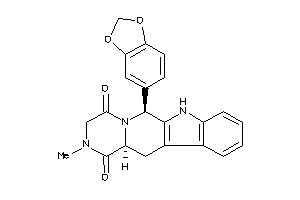
-
8204642
-
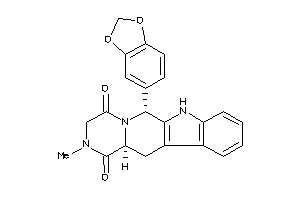
-
538404
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNCG-1-E |
Phosphodiesterase 6H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
CNRG-1-E |
Phosphodiesterase 6G (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE11-1-E |
Phosphodiesterase 11A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
PDE6A-1-E |
Rod CGMP-specific 3',5'-cyclic Phosphodiesterase Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6B-1-E |
Phosphodiesterase 6B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6C-1-E |
Phosphodiesterase 6C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.29 |
Binding ≤ 10μM
|
|
PDE6D-1-E |
Phosphodiesterase 6D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.26 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
500 |
0.30 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE11_HUMAN |
Q9HCR9
|
Phosphodiesterase 11A, Human |
290 |
0.32 |
Binding ≤ 1μM
|
|
PDE5A_HUMAN |
O76074
|
Phosphodiesterase 5A, Human |
1.3 |
0.43 |
Binding ≤ 1μM
|
|
PDE5A_RAT |
O54735
|
Phosphodiesterase 5A, Rat |
5 |
0.40 |
Binding ≤ 1μM
|
|
PDE5A_BOVIN |
Q28156
|
Phosphodiesterase 5A, Bovin |
12 |
0.38 |
Binding ≤ 1μM
|
|
PDE6A_HUMAN |
P16499
|
Phosphodiesterase 6A, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
PDE6B_HUMAN |
P35913
|
Phosphodiesterase 6B, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
PDE6C_HUMAN |
P51160
|
Phosphodiesterase 6C, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
PDE6D_HUMAN |
O43924
|
Phosphodiesterase 6D, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
CNRG_HUMAN |
P18545
|
Phosphodiesterase 6G, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
CNCG_HUMAN |
Q13956
|
Phosphodiesterase 6H, Human |
980 |
0.29 |
Binding ≤ 1μM
|
|
PDE11_HUMAN |
Q9HCR9
|
Phosphodiesterase 11A, Human |
290 |
0.32 |
Binding ≤ 10μM
|
|
PDE5A_BOVIN |
Q28156
|
Phosphodiesterase 5A, Bovin |
12 |
0.38 |
Binding ≤ 10μM
|
|
PDE5A_HUMAN |
O76074
|
Phosphodiesterase 5A, Human |
1.3 |
0.43 |
Binding ≤ 10μM
|
|
PDE5A_RAT |
O54735
|
Phosphodiesterase 5A, Rat |
5 |
0.40 |
Binding ≤ 10μM
|
|
PDE6A_HUMAN |
P16499
|
Phosphodiesterase 6A, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
PDE6A_BOVIN |
P11541
|
Phosphodiesterase 6A, Bovin |
3000 |
0.27 |
Binding ≤ 10μM
|
|
PDE6B_HUMAN |
P35913
|
Phosphodiesterase 6B, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
PDE6B_BOVIN |
P23439
|
Phosphodiesterase 6B, Bovin |
5100 |
0.26 |
Binding ≤ 10μM
|
|
PDE6C_BOVIN |
P16586
|
Phosphodiesterase 6C, Bovin |
5100 |
0.26 |
Binding ≤ 10μM
|
|
PDE6C_HUMAN |
P51160
|
Phosphodiesterase 6C, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
PDE6D_BOVIN |
Q95142
|
Phosphodiesterase 6D, Bovin |
5100 |
0.26 |
Binding ≤ 10μM
|
|
PDE6D_HUMAN |
O43924
|
Phosphodiesterase 6D, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
CNRG_HUMAN |
P18545
|
Phosphodiesterase 6G, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
CNRG_BOVIN |
P04972
|
Phosphodiesterase 6G, Bovin |
5100 |
0.26 |
Binding ≤ 10μM
|
|
CNCG_BOVIN |
P22571
|
Phosphodiesterase 6H, Bovin |
5100 |
0.26 |
Binding ≤ 10μM
|
|
CNCG_HUMAN |
Q13956
|
Phosphodiesterase 6H, Human |
1260 |
0.28 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
150 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
6.81 |
-13.33 |
1 |
7 |
0 |
75 |
389.411 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
3947571
-
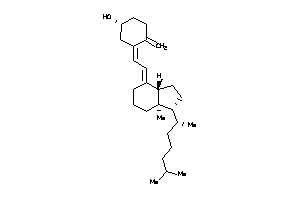
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-1-E |
Dual Specificity Phosphatase Cdc25A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.53 |
-2.24 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.76 |
10.23 |
-3.75 |
2 |
2 |
0 |
40 |
400.647 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
34337995
-
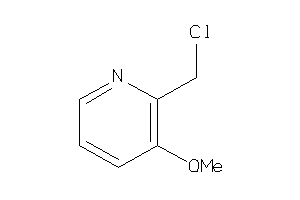
-
39038289
-
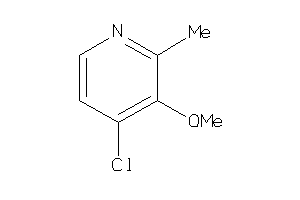
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
3.82 |
-4.13 |
0 |
2 |
0 |
22 |
192.045 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
2.34 |
-14.6 |
0 |
4 |
0 |
56 |
280.082 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
9.62 |
-14.14 |
0 |
5 |
0 |
61 |
310.712 |
4 |
↓
|
|
|
|
Analogs
-
39280667
-
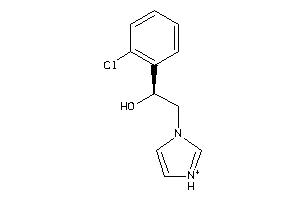
-
39280669
-
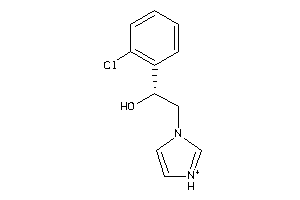
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
6.83 |
-36.08 |
2 |
3 |
1 |
39 |
258.128 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.09 |
6.32 |
-9.49 |
1 |
3 |
0 |
38 |
257.12 |
3 |
↓
|
|
|
|
Analogs
-
39280667
-

-
39280669
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
6.87 |
-34.37 |
2 |
3 |
1 |
39 |
258.128 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
1.82 |
-14.93 |
0 |
3 |
0 |
34 |
255.104 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.11 |
2.11 |
-41.34 |
1 |
3 |
1 |
36 |
256.112 |
3 |
↓
|
|
|
|
Analogs
-
4241109
-
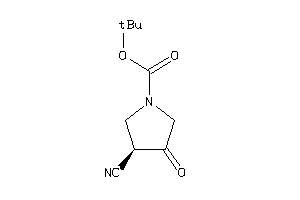
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
3.96 |
-12.01 |
0 |
5 |
0 |
70 |
210.233 |
2 |
↓
|
|
|
|
Analogs
-
4241114
-
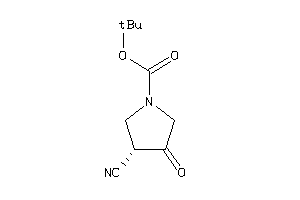
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
3.98 |
-10.5 |
0 |
5 |
0 |
70 |
210.233 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
6.23 |
-12.54 |
0 |
4 |
0 |
60 |
358.85 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-0.72 |
-3.01 |
2 |
1 |
0 |
26 |
237.015 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
-0.53 |
-6.22 |
0 |
4 |
0 |
44 |
259.092 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
5.44 |
-5.98 |
0 |
2 |
0 |
26 |
176.215 |
1 |
↓
|
|
|
|
Analogs
-
34359996
-
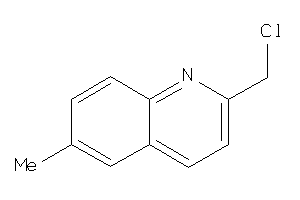
-
39321580
-
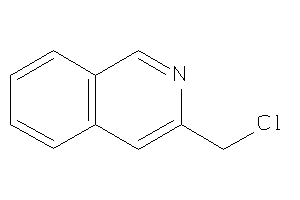
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
0.15 |
-6.98 |
0 |
1 |
0 |
13 |
177.634 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
0.2 |
-28.19 |
1 |
1 |
1 |
14 |
178.642 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
7.71 |
-20.94 |
0 |
4 |
0 |
48 |
223.182 |
3 |
↓
|
|
|
|
Analogs
-
3944782
-
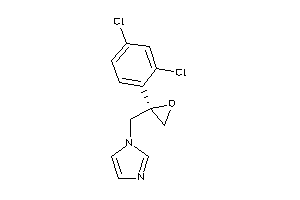
-
596881
-
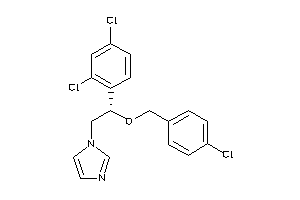
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
5 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
GRM6-2-E |
Metabotropic Glutamate Receptor 6 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
851 |
0.34 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
2 |
0.49 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.37 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
330 |
0.36 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 1μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 1μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 10μM
|
|
Z50416 |
Z50416
|
Aspergillus Fumigatus |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442 |
Z50442
|
Candida Albicans |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50443 |
Z50443
|
Candida Glabrata |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50408 |
Z50408
|
Issatchenkia Orientalis |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409 |
Z50409
|
Kluyveromyces Marxianus |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50459 |
Z50459
|
Leishmania Donovani |
6000 |
0.29 |
Functional ≤ 10μM
|
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
7800 |
0.29 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
2.03 |
0.49 |
Functional ≤ 10μM
|
|
Z102121 |
Z102121
|
Trichophyton Mentagrophytes |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50046 |
Z50046
|
Trichophyton Quinckeanum |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50452 |
Z50452
|
Trichophyton Rubrum |
140 |
0.38 |
Functional ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
180 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
13.6 |
-35.01 |
1 |
3 |
1 |
28 |
417.143 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
13.09 |
-6.46 |
0 |
3 |
0 |
27 |
416.135 |
6 |
↓
|
|
|
|
Analogs
-
643055
-
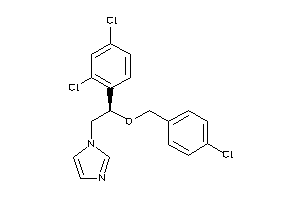
-
896740
-
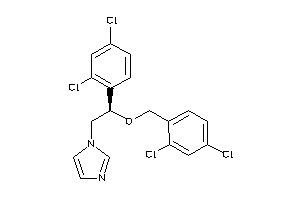
-
3872945
-
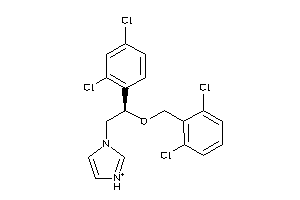
-
3944782
-

-
596881
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
5 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
GRM6-2-E |
Metabotropic Glutamate Receptor 6 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
851 |
0.34 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
2 |
0.49 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.37 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
330 |
0.36 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 1μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 1μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 10μM
|
|
Z50416 |
Z50416
|
Aspergillus Fumigatus |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442 |
Z50442
|
Candida Albicans |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50443 |
Z50443
|
Candida Glabrata |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50408 |
Z50408
|
Issatchenkia Orientalis |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409 |
Z50409
|
Kluyveromyces Marxianus |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50459 |
Z50459
|
Leishmania Donovani |
6000 |
0.29 |
Functional ≤ 10μM
|
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
7800 |
0.29 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
2.03 |
0.49 |
Functional ≤ 10μM
|
|
Z102121 |
Z102121
|
Trichophyton Mentagrophytes |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50046 |
Z50046
|
Trichophyton Quinckeanum |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50452 |
Z50452
|
Trichophyton Rubrum |
140 |
0.38 |
Functional ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
180 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.15 |
-35.65 |
1 |
3 |
1 |
28 |
417.143 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
13.63 |
-6.4 |
0 |
3 |
0 |
27 |
416.135 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
-0.07 |
-3.28 |
0 |
2 |
0 |
26 |
166.97 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2070 |
0.26 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.24 |
Binding ≤ 10μM |
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.28 |
Functional ≤ 10μM
|
|
Z80006-1-O |
3T3-L1 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
7.5 |
-12.36 |
2 |
6 |
0 |
85 |
441.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2070 |
0.26 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.24 |
Binding ≤ 10μM |
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.28 |
Functional ≤ 10μM
|
|
Z80006-1-O |
3T3-L1 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
7.29 |
-12.45 |
2 |
6 |
0 |
85 |
441.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2070 |
0.26 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.24 |
Binding ≤ 10μM |
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.28 |
Functional ≤ 10μM
|
|
Z80006-1-O |
3T3-L1 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
7.5 |
-12.94 |
2 |
6 |
0 |
85 |
441.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2070 |
0.26 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.24 |
Binding ≤ 10μM |
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.28 |
Functional ≤ 10μM
|
|
Z80006-1-O |
3T3-L1 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
7.3 |
-12.93 |
2 |
6 |
0 |
85 |
441.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
1.76 |
-5.23 |
1 |
1 |
0 |
20 |
130.093 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
1.82 |
-5.33 |
1 |
1 |
0 |
20 |
148.083 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
1.78 |
-5.91 |
1 |
1 |
0 |
20 |
130.093 |
0 |
↓
|
|
|
|
Analogs
-
34563029
-
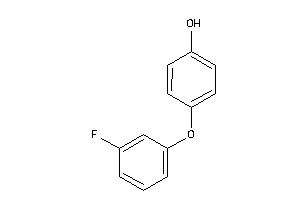
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
3.87 |
-3.5 |
0 |
1 |
0 |
9 |
126.13 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
4.05 |
-5.96 |
0 |
1 |
0 |
9 |
144.12 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
5.61 |
-3.18 |
0 |
1 |
0 |
17 |
276.563 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
2.08 |
-7.65 |
1 |
2 |
0 |
44 |
137.113 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
4.92 |
-7.31 |
0 |
1 |
0 |
24 |
121.114 |
0 |
↓
|
|
|
|
Analogs
-
164883
-
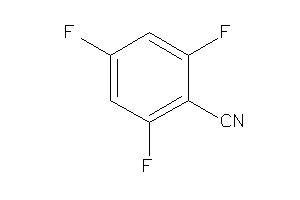
-
39278278
-
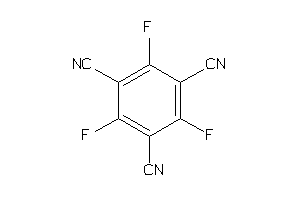
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
4.98 |
-5.71 |
0 |
1 |
0 |
24 |
139.104 |
0 |
↓
|
|
|
|
Analogs
-
39128310
-
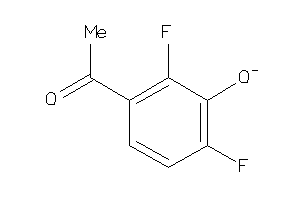
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
3.16 |
-13.42 |
0 |
2 |
0 |
26 |
186.157 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
2.75 |
-5.97 |
2 |
2 |
0 |
35 |
191.152 |
2 |
↓
|
|
|
|
Analogs
-
6091910
-
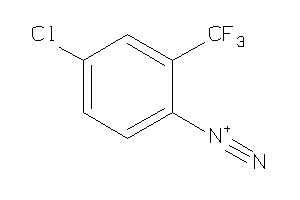
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
1.31 |
-3.13 |
2 |
1 |
0 |
26 |
195.571 |
1 |
↓
|
|
|
|
Analogs
-
34974896
-
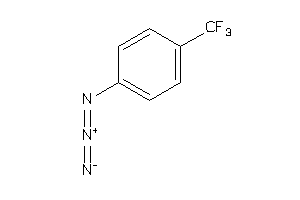
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
3.37 |
-3.6 |
2 |
1 |
0 |
26 |
161.126 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
39051303
-
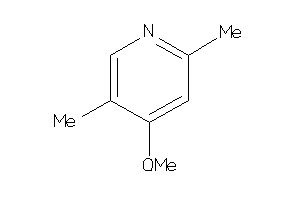
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
1.15 |
-5.2 |
0 |
2 |
0 |
22 |
185.654 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.54 |
1.21 |
-30.45 |
1 |
2 |
1 |
23 |
186.662 |
2 |
↓
|
|
|
|
Analogs
-
33787715
-
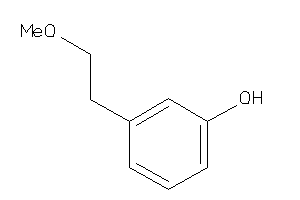
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
2.15 |
-4.28 |
1 |
2 |
0 |
29 |
152.193 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
-1.76 |
-3.42 |
0 |
4 |
0 |
43 |
184.239 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
8.77 |
-8.47 |
0 |
1 |
0 |
17 |
208.26 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
8.79 |
-6.91 |
0 |
2 |
0 |
26 |
243.693 |
2 |
↓
|
|