|
|
Analogs
-
3831613
-
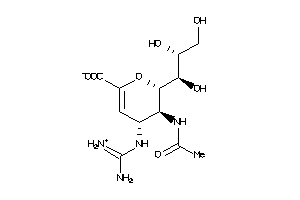
-
3831612
-
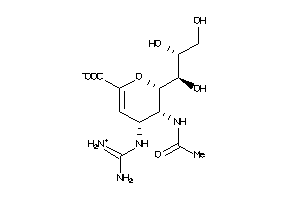
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
3 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.51 |
-12.29 |
-65.75 |
9 |
11 |
0 |
202 |
332.313 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.59 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
840 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
-0.99 |
-41.7 |
3 |
3 |
1 |
41 |
205.281 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
3.13 |
-9.55 |
3 |
5 |
0 |
78 |
217.228 |
2 |
↓
|
|
|
|
Analogs
-
44512272
-

-
44512280
-
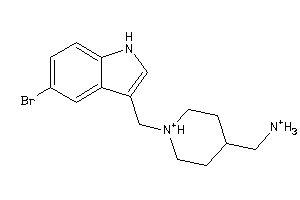
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.43 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
570 |
0.51 |
Binding ≤ 10μM
|
|
DRD2-23-E |
Dopamine D2 Receptor (cluster #23 Of 24), Eukaryotic |
Eukaryotes |
5500 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
0.66 |
-35.2 |
2 |
2 |
1 |
20 |
229.347 |
2 |
↓
|
|
|
|
Analogs
-
4090202
-
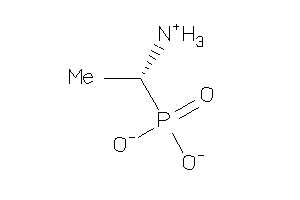
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.18 |
-3.38 |
-75.24 |
3 |
4 |
-1 |
90 |
124.056 |
1 |
↓
|
|
|
|
Analogs
-
5157947
-
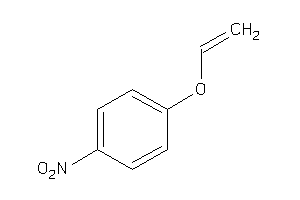
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
4.46 |
-6.55 |
0 |
4 |
0 |
55 |
153.137 |
2 |
↓
|
|
|
|
Analogs
-
44512272
-

-
44512280
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
8.34 |
-34.55 |
2 |
2 |
1 |
20 |
215.32 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.80 |
6.12 |
-4.64 |
1 |
2 |
0 |
19 |
214.312 |
2 |
↓
|
|
|
|
Analogs
-
38522503
-
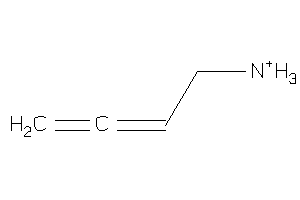
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.08 |
-38.6 |
3 |
1 |
1 |
28 |
58.104 |
1 |
↓
|
|
|
|
Analogs
-
5344554
-
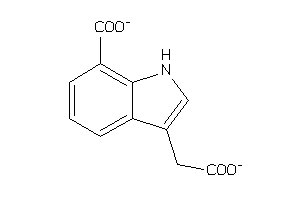
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
4.57 |
-52.71 |
1 |
4 |
-1 |
73 |
188.162 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
2.97 |
-48.95 |
1 |
5 |
-1 |
86 |
242.276 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
34007650
-
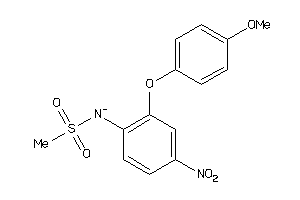
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3760 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-8-E |
Cyclooxygenase-2 (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
400 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
4.93 |
-37.19 |
0 |
7 |
-1 |
103 |
307.307 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.81 |
4.82 |
-11.73 |
1 |
7 |
0 |
101 |
308.315 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO-1-E |
Dopamine Beta-hydroxylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.62 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO_BOVIN |
P15101
|
Dopamine Beta-hydroxylase, Bovin |
5300 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
6.63 |
-16.82 |
1 |
2 |
0 |
21 |
196.3 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.85 |
0.05 |
-30.09 |
1 |
2 |
1 |
19 |
197.308 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO-1-E |
Dopamine Beta-hydroxylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.56 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO_BOVIN |
P15101
|
Dopamine Beta-hydroxylase, Bovin |
6000 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
7.81 |
-20.08 |
1 |
2 |
0 |
21 |
210.327 |
2 |
↓
|
|
|
|
Analogs
-
12503190
-
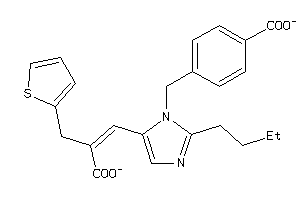
-
29319828
-
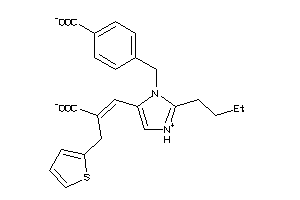
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
12.07 |
-108.21 |
1 |
6 |
-1 |
99 |
409.487 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.33 |
0.54 |
-105.97 |
1 |
6 |
-1 |
95 |
409.487 |
9 |
↓
|
|
|
|
Analogs
-
25492647
-
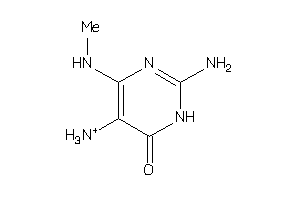
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
0.58 |
-12.64 |
4 |
5 |
0 |
84 |
140.146 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
38413756
-
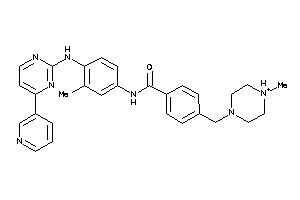
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199-1-E |
BCR/ABL P210 Fusion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.23 |
Binding ≤ 10μM
|
|
ABCG2-2-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4898 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.24 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
468 |
0.24 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
30 |
0.28 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
76 |
0.27 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.20 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.20 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.23 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.28 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.30 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.19 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.21 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.27 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.25 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.27 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.19 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.22 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.20 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Functional ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.19 |
Functional ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
9100 |
0.19 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
6 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
10000 |
0.19 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199_HUMAN |
A1Z199
|
BCR/ABL P210 Fusion Protein, Human |
204 |
0.25 |
Binding ≤ 1μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
1.1 |
0.34 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.27 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
31.9 |
0.28 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
30.2 |
0.28 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
528 |
0.24 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
75.7 |
0.27 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
392 |
0.24 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
109 |
0.26 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
980 |
0.23 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
468 |
0.24 |
Binding ≤ 1μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
141 |
0.26 |
Binding ≤ 1μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
0.7 |
0.35 |
Binding ≤ 1μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
19 |
0.29 |
Binding ≤ 1μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
160 |
0.26 |
Binding ≤ 1μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
14 |
0.30 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
1000 |
0.23 |
Binding ≤ 1μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
100 |
0.26 |
Binding ≤ 1μM
|
|
ABL1_MOUSE |
P00520
|
Tyrosine-protein Kinase ABL, Mouse |
10.8 |
0.30 |
Binding ≤ 1μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
430 |
0.24 |
Binding ≤ 1μM
|
|
ABL2_HUMAN |
P42684
|
Tyrosine-protein Kinase ABL2, Human |
10 |
0.30 |
Binding ≤ 1μM
|
|
BLK_HUMAN |
P51451
|
Tyrosine-protein Kinase BLK, Human |
520 |
0.24 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
160 |
0.26 |
Binding ≤ 1μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
220 |
0.25 |
Binding ≤ 1μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
3380 |
0.21 |
Binding ≤ 10μM
|
|
A1Z199_HUMAN |
A1Z199
|
BCR/ABL P210 Fusion Protein, Human |
204 |
0.25 |
Binding ≤ 10μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
1.1 |
0.34 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
3200 |
0.21 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
3100 |
0.21 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
31.9 |
0.28 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
30.2 |
0.28 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
75.7 |
0.27 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
468 |
0.24 |
Binding ≤ 10μM
|
|
CDK19_HUMAN |
Q9BWU1
|
Cell Division Cycle 2-like Protein Kinase 6, Human |
5500 |
0.20 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
141 |
0.26 |
Binding ≤ 10μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
2100 |
0.21 |
Binding ≤ 10μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
4500 |
0.20 |
Binding ≤ 10μM
|
|
EPHA8_HUMAN |
P29322
|
Ephrin Type-A Receptor 8, Human |
1400 |
0.22 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
7600 |
0.19 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
0.7 |
0.35 |
Binding ≤ 10μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
1274 |
0.22 |
Binding ≤ 10μM
|
|
MELK_HUMAN |
Q14680
|
Maternal Embryonic Leucine Zipper Kinase, Human |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MLTK_HUMAN |
Q9NYL2
|
Mixed Lineage Kinase 7, Human |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
160 |
0.26 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
14 |
0.30 |
Binding ≤ 10μM
|
|
ST17A_HUMAN |
Q9UEE5
|
Serine/threonine-protein Kinase 17A, Human |
2800 |
0.21 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
3300 |
0.21 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
1000 |
0.23 |
Binding ≤ 10μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
7800 |
0.19 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
1700 |
0.22 |
Binding ≤ 10μM
|
|
TNI3K_HUMAN |
Q59H18
|
Serine/threonine-protein Kinase TNNI3K, Human |
4300 |
0.20 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
100 |
0.26 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
430 |
0.24 |
Binding ≤ 10μM
|
|
ABL1_MOUSE |
P00520
|
Tyrosine-protein Kinase ABL, Mouse |
10.8 |
0.30 |
Binding ≤ 10μM
|
|
ABL2_HUMAN |
P42684
|
Tyrosine-protein Kinase ABL2, Human |
10 |
0.30 |
Binding ≤ 10μM
|
|
BLK_HUMAN |
P51451
|
Tyrosine-protein Kinase BLK, Human |
520 |
0.24 |
Binding ≤ 10μM
|
|
FGR_HUMAN |
P09769
|
Tyrosine-protein Kinase FGR, Human |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
1500 |
0.22 |
Binding ≤ 10μM
|
|
FYN_HUMAN |
P06241
|
Tyrosine-protein Kinase FYN, Human |
3100 |
0.21 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
160 |
0.26 |
Binding ≤ 10μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
220 |
0.25 |
Binding ≤ 10μM
|
|
KSYK_HUMAN |
P43405
|
Tyrosine-protein Kinase SYK, Human |
5000 |
0.20 |
Binding ≤ 10μM
|
|
Z80036 |
Z80036
|
BaF3 (IL-3-dependent Pro-B-cells) |
400 |
0.24 |
Functional ≤ 10μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
90 |
0.27 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
10000 |
0.19 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
90 |
0.27 |
Functional ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
96 |
0.27 |
Functional ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
219 |
0.25 |
Functional ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
9000 |
0.19 |
Functional ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
90 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
13.15 |
-49.4 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
13.03 |
-52.36 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
10.78 |
-14.7 |
2 |
8 |
0 |
86 |
493.615 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.65 |
2.45 |
-19.35 |
4 |
8 |
0 |
130 |
185.143 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
47 |
0.41 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
3.79 |
-50.99 |
2 |
6 |
1 |
57 |
347.435 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.25 |
2.37 |
-10.67 |
1 |
6 |
0 |
52 |
346.427 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
275 |
0.37 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
3.77 |
-49.25 |
2 |
6 |
1 |
57 |
347.435 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.25 |
2.4 |
-10.91 |
1 |
6 |
0 |
52 |
346.427 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
34530759
-
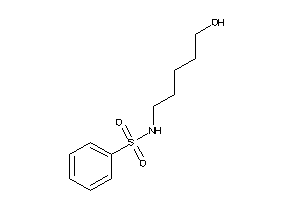
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
3.75 |
-49.73 |
1 |
5 |
-1 |
86 |
256.303 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.30 |
1.77 |
-13.71 |
2 |
5 |
0 |
83 |
257.311 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
5.31 |
-51.06 |
1 |
5 |
-1 |
86 |
284.357 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.31 |
3.34 |
-13.67 |
2 |
5 |
0 |
83 |
285.365 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO-1-E |
Dopamine Beta-hydroxylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.61 |
-20.15 |
1 |
2 |
0 |
21 |
230.745 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
3.04 |
-47.62 |
0 |
2 |
-1 |
40 |
99.109 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
5371008
-
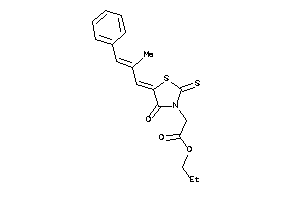
-
5371009
-
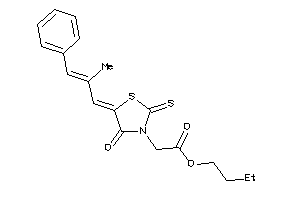
-
5371011
-

-
5376115
-
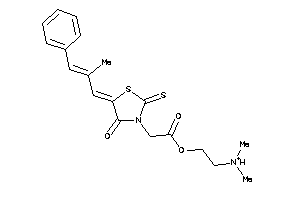
-
13507152
-
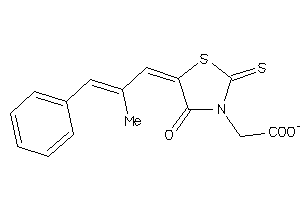
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1A1-1-E |
Aldehyde Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.39 |
Binding ≤ 10μM
|
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.43 |
Binding ≤ 10μM
|
|
ALDR-2-E |
Aldose Reductase (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
170 |
0.45 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1BA_HUMAN |
O60218
|
Aldo-keto Reductase Family 1 Member B10, Human |
330 |
0.43 |
Binding ≤ 1μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
170 |
0.45 |
Binding ≤ 1μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
10 |
0.53 |
Binding ≤ 1μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
10 |
0.53 |
Binding ≤ 1μM
|
|
AK1A1_HUMAN |
P14550
|
Aldehyde Reductase, Human |
2600 |
0.37 |
Binding ≤ 10μM
|
|
AK1A1_RAT |
P51635
|
Aldehyde Reductase, Rat |
1500 |
0.39 |
Binding ≤ 10μM
|
|
AK1A1_PIG |
P50578
|
Aldehyde Reductase, Pig |
1500 |
0.39 |
Binding ≤ 10μM
|
|
AK1BA_HUMAN |
O60218
|
Aldo-keto Reductase Family 1 Member B10, Human |
330 |
0.43 |
Binding ≤ 10μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
170 |
0.45 |
Binding ≤ 10μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
10 |
0.53 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
10 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
9.92 |
-56.16 |
0 |
4 |
-1 |
62 |
318.399 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
9.72 |
-42.06 |
2 |
7 |
1 |
72 |
427.447 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
7.51 |
-16.59 |
1 |
7 |
0 |
71 |
426.439 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
5.23 |
-34.59 |
1 |
5 |
1 |
47 |
264.349 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.23 |
7.32 |
-43.39 |
1 |
5 |
1 |
47 |
264.349 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.23 |
4.95 |
-8.51 |
0 |
5 |
0 |
46 |
263.341 |
6 |
↓
|
|
|
|
Analogs
-
4899421
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.33 |
-2.73 |
-64.96 |
4 |
5 |
0 |
96 |
312.369 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.63 |
12 |
-39.39 |
3 |
1 |
1 |
28 |
270.525 |
16 |
↓
|
|
|
|
Analogs
-
32155422
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
0.66 |
-10.6 |
3 |
4 |
0 |
72 |
111.104 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.35 |
Binding ≤ 10μM
|
|
DYR-2-E |
Dihydrofolate Reductase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
1600 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
12.08 |
-35.97 |
5 |
8 |
1 |
128 |
379.444 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.55 |
11.62 |
-9.15 |
4 |
8 |
0 |
127 |
378.436 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
6.53 |
-39.03 |
9 |
8 |
1 |
148 |
447.563 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.33 |
6.63 |
-43.48 |
9 |
8 |
1 |
150 |
447.563 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.33 |
6.9 |
-102.54 |
10 |
8 |
2 |
152 |
448.571 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
3.98 |
-53.54 |
5 |
9 |
-1 |
153 |
425.465 |
11 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO-1-E |
Dopamine Beta-hydroxylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6300 |
0.61 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DOPO_BOVIN |
P15101
|
Dopamine Beta-hydroxylase, Bovin |
6300 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
5.85 |
-20.86 |
1 |
3 |
0 |
34 |
180.232 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.18 |
1.09 |
-30.75 |
1 |
3 |
1 |
32 |
181.24 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
5.78 |
-21.32 |
3 |
4 |
0 |
67 |
251.289 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.04 |
1.39 |
-45.73 |
0 |
7 |
-1 |
101 |
295.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-5.23 |
-81.82 |
6 |
7 |
0 |
139 |
259.306 |
8 |
↓
|
|