|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.73 |
-6.16 |
-40.15 |
6 |
9 |
1 |
134 |
333.361 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.73 |
-7.31 |
-11.66 |
5 |
9 |
0 |
130 |
332.353 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.73 |
-4.87 |
-109.15 |
7 |
9 |
2 |
139 |
334.369 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
0.92 |
-16.47 |
2 |
7 |
0 |
106 |
310.302 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2360 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.29 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
106 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1585 |
0.29 |
Binding ≤ 10μM
|
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
1 |
0.45 |
Binding ≤ 10μM
|
|
Z50597-5-O |
Rattus Norvegicus (cluster #5 Of 5), Other |
Other |
3000 |
0.28 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.86 |
0.45 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
0.1 |
0.50 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.254 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
0.27 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.17 |
0.49 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
0.19 |
0.49 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.21 |
0.48 |
Binding ≤ 1μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.83 |
0.45 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
106 |
0.35 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
MT3_HUMAN |
P25713
|
Metallothionein-3, Human |
7.8 |
0.41 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
0.55 |
0.46 |
Binding ≤ 1μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
0.33 |
0.47 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.86 |
0.45 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
0.1 |
0.50 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
0.27 |
0.48 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.17 |
0.49 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.254 |
0.48 |
Binding ≤ 10μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.83 |
0.45 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
0.19 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.21 |
0.48 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
ADA2A_PIG |
P18871
|
Alpha-2a Adrenergic Receptor, Pig |
4100 |
0.27 |
Binding ≤ 10μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
106 |
0.35 |
Binding ≤ 10μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
8300 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1584.89319 |
0.29 |
Binding ≤ 10μM
|
|
MT3_HUMAN |
P25713
|
Metallothionein-3, Human |
7.8 |
0.41 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
3000 |
0.28 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
2360 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
0.55 |
0.46 |
Binding ≤ 10μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
0.33 |
0.47 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1500 |
0.29 |
Binding ≤ 10μM
|
|
S22A1_HUMAN |
O15245
|
Solute Carrier Family 22 Member 1, Human |
9900 |
0.25 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.25 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
0.13 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
7.64 |
-36.73 |
3 |
9 |
1 |
108 |
384.416 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
7.26 |
-15.54 |
2 |
9 |
0 |
107 |
383.408 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
0.64 |
-39 |
4 |
2 |
1 |
48 |
152.217 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.61 |
-0.18 |
-4.33 |
3 |
2 |
0 |
46 |
151.209 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
0.63 |
-39.11 |
4 |
2 |
1 |
48 |
152.217 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.61 |
-0.08 |
-3.84 |
3 |
2 |
0 |
46 |
151.209 |
2 |
↓
|
|
|
|
Analogs
-
16248873
-
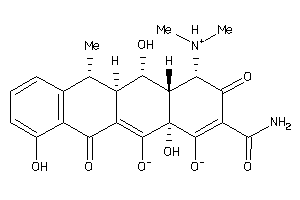
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
0.98 |
-89.55 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.56 |
-4.34 |
-53.29 |
6 |
10 |
-1 |
185 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-2.24 |
-69.99 |
6 |
10 |
-1 |
184 |
443.432 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
9.27 |
-20.79 |
1 |
8 |
0 |
110 |
346.339 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
3.81 |
-8.91 |
2 |
6 |
0 |
79 |
249.27 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.54 |
3.8 |
-10.36 |
2 |
6 |
0 |
79 |
249.27 |
4 |
↓
|
|
|
|
Analogs
-
5784598
-
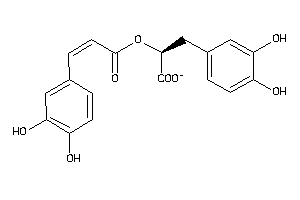
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
2.27 |
-59.27 |
4 |
8 |
-1 |
147 |
359.31 |
7 |
↓
|
|
|
|
Analogs
-
1481729
-
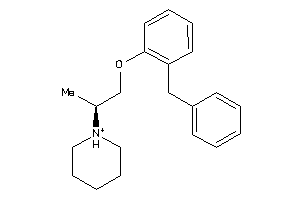
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
2.35 |
-33.5 |
1 |
2 |
1 |
13 |
310.461 |
6 |
↓
|
|
|
|
Analogs
-
3635554
-
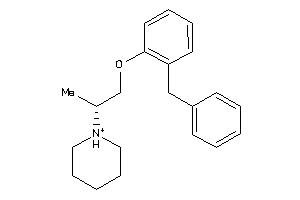
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
2.7 |
-34.22 |
1 |
2 |
1 |
13 |
310.461 |
6 |
↓
|
|
|
|
Analogs
-
37867490
-

-
37874880
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.10 |
-10.88 |
-76.16 |
11 |
12 |
1 |
227 |
380.374 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.10 |
-10.24 |
-25.26 |
10 |
12 |
0 |
226 |
379.366 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.10 |
-11.21 |
-34.53 |
10 |
12 |
0 |
226 |
379.366 |
4 |
↓
|
|
|
|
Analogs
-
8551990
-
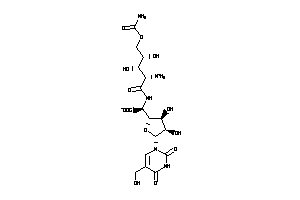
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.59 |
-15.6 |
-79.56 |
12 |
18 |
0 |
314 |
507.409 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.59 |
-15.21 |
-64.04 |
11 |
18 |
-1 |
313 |
506.401 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.59 |
-14.52 |
-75.37 |
12 |
18 |
0 |
314 |
507.409 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.59 |
-14.69 |
-61.97 |
11 |
18 |
-1 |
313 |
506.401 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.59 |
-13.81 |
-86.14 |
12 |
18 |
0 |
314 |
507.409 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.59 |
-13.99 |
-70.49 |
11 |
18 |
-1 |
313 |
506.401 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.59 |
-13.97 |
-76.45 |
12 |
18 |
0 |
314 |
507.409 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.59 |
-14.17 |
-62 |
11 |
18 |
-1 |
313 |
506.401 |
11 |
↓
|
|
|
|
Analogs
-
32143440
-

-
39383244
-
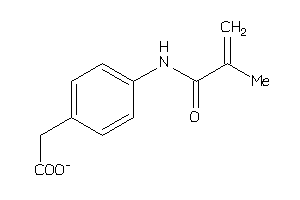
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
4.74 |
-53.38 |
1 |
4 |
-1 |
69 |
192.194 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.27 |
-7.26 |
-15.96 |
7 |
10 |
0 |
170 |
309.282 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.27 |
-7.32 |
-28.07 |
8 |
10 |
1 |
172 |
310.29 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.27 |
-6.48 |
-15.28 |
7 |
10 |
0 |
170 |
309.282 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.27 |
-6.54 |
-28.71 |
8 |
10 |
1 |
172 |
310.29 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.27 |
-5.96 |
-17.17 |
7 |
10 |
0 |
170 |
309.282 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.27 |
-6.02 |
-30.58 |
8 |
10 |
1 |
172 |
310.29 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.27 |
-6.28 |
-16.31 |
7 |
10 |
0 |
170 |
309.282 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.27 |
-6.34 |
-27.81 |
8 |
10 |
1 |
172 |
310.29 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
8.71 |
-50.57 |
2 |
6 |
1 |
63 |
402.902 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.41 |
6.44 |
-14.09 |
1 |
6 |
0 |
62 |
401.894 |
6 |
↓
|
|
|
|
Analogs
-
169128
-
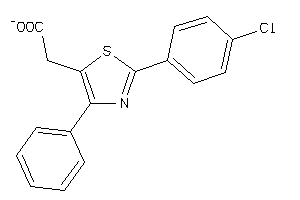
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.40 |
Binding ≤ 10μM
|
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.80 |
10.81 |
-47.84 |
0 |
3 |
-1 |
53 |
328.8 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6100 |
0.61 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.61 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.83 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.83 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.83 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
6100 |
0.61 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6000 |
0.61 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
2000 |
0.66 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
2500 |
0.65 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
2500 |
0.65 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
270 |
0.77 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
390 |
0.75 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
6000 |
0.61 |
Functional ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
6000 |
0.61 |
Functional ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
6000 |
0.61 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
6000 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.41 |
-0.72 |
-41.71 |
4 |
3 |
1 |
57 |
168.216 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.41 |
-2.17 |
-5.18 |
3 |
3 |
0 |
52 |
167.208 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.13 |
4.89 |
-11.19 |
1 |
6 |
0 |
84 |
219.628 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.13 |
4.3 |
-12.26 |
1 |
6 |
0 |
84 |
219.628 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
3.64 |
-46.51 |
5 |
3 |
1 |
63 |
379.116 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.74 |
2.42 |
-3.86 |
4 |
3 |
0 |
58 |
378.108 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.58 |
8.6 |
-2.12 |
0 |
0 |
0 |
0 |
194.343 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.57 |
-2.59 |
-47.5 |
5 |
3 |
1 |
68 |
154.189 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.57 |
-2.98 |
-5.66 |
4 |
3 |
0 |
66 |
153.181 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.57 |
-2.68 |
-46.85 |
5 |
3 |
1 |
68 |
154.189 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.57 |
-3.13 |
-5.54 |
4 |
3 |
0 |
66 |
153.181 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
330
-
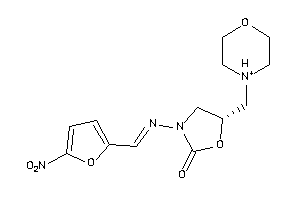
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.69 |
1.78 |
-33.9 |
0 |
10 |
0 |
113 |
324.293 |
5 |
↓
|
|
|
|
Analogs
-
330
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.69 |
1.68 |
-34.32 |
0 |
10 |
0 |
113 |
324.293 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
5.8 |
-8.94 |
2 |
5 |
0 |
70 |
247.298 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.47 |
5.8 |
-9.29 |
2 |
5 |
0 |
70 |
247.298 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
32221120
-

-
32221121
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
9.16 |
-36.71 |
1 |
2 |
1 |
14 |
254.353 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.42 |
6.93 |
-118.09 |
2 |
6 |
-2 |
123 |
263.249 |
6 |
↓
|
|
|
|
Analogs
-
32221120
-

-
32221121
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
9.07 |
-36.58 |
1 |
2 |
1 |
14 |
254.353 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
7.52 |
-17.36 |
2 |
6 |
0 |
96 |
293.319 |
8 |
↓
|
|
|
|
Analogs
-
4072734
-
-
4072735
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3217 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
3217 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
13.51 |
-42.06 |
2 |
1 |
1 |
17 |
316.468 |
7 |
↓
|
|
|
|
Analogs
-
4072734
-

-
4072735
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3217 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
3217 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
13.5 |
-42.05 |
2 |
1 |
1 |
17 |
316.468 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 1μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
0.52 |
0.72 |
Binding ≤ 1μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
0.4 |
0.73 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
110 |
0.54 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
0.52 |
0.72 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
0.4 |
0.73 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
110 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
2600 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.18 |
-42.7 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
1.9 |
-9.07 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
0.52 |
0.72 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 1μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
0.4 |
0.73 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
110 |
0.54 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
100 |
0.54 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
0.52 |
0.72 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
0.4 |
0.73 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1 |
0.70 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
110 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
2600 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.19 |
-42.74 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
2.01 |
-8.94 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.87 |
2.82 |
-43.8 |
2 |
5 |
1 |
48 |
267.349 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.87 |
1.46 |
-7.3 |
1 |
5 |
0 |
43 |
266.341 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
8500 |
0.59 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
873 |
0.71 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
7330 |
0.60 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
8690 |
0.59 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
83 |
0.83 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
508 |
0.73 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
41 |
0.86 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
3200 |
0.64 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
970 |
0.70 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
4000 |
0.63 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
2800 |
0.65 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
920 |
0.70 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
920 |
0.70 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
4800 |
0.62 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
75 |
0.83 |
Binding ≤ 10μM
|
|
CAH9-9-E |
Carbonic Anhydrase IX (cluster #9 Of 11), Eukaryotic |
Eukaryotes |
103 |
0.82 |
Binding ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
433 |
0.74 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
170 |
0.79 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
82.5 |
0.83 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
508 |
0.73 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
170 |
0.79 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
433 |
0.74 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
350 |
0.75 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
830 |
0.71 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
970 |
0.70 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
170 |
0.79 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
103 |
0.82 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
920 |
0.70 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
920 |
0.70 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
75 |
0.83 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.3 |
1.11 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
41 |
0.86 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
82.5 |
0.83 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
433 |
0.74 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
350 |
0.75 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
1109 |
0.69 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
508 |
0.73 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
830 |
0.71 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
970 |
0.70 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
170 |
0.79 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
2800 |
0.65 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
2800 |
0.65 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
103 |
0.82 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
920 |
0.70 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
920 |
0.70 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
4800 |
0.62 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
75 |
0.83 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.3 |
1.11 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
41 |
0.86 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
3200 |
0.64 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
7330 |
0.60 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
8690 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
-2.51 |
-57.02 |
5 |
4 |
1 |
88 |
187.244 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.18 |
-2.91 |
-10.08 |
4 |
4 |
0 |
86 |
186.236 |
2 |
↓
|
|