|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
207 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
0.19 |
-9.4 |
1 |
3 |
0 |
46 |
300.398 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
34610163
-
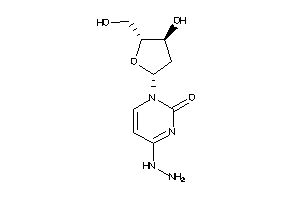
-
4253
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
-5.1 |
-22.79 |
4 |
7 |
0 |
111 |
227.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-1.85 |
-13.19 |
2 |
4 |
0 |
74 |
346.467 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
4228257
-
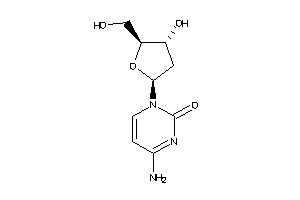
-
4228258
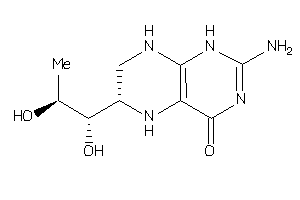
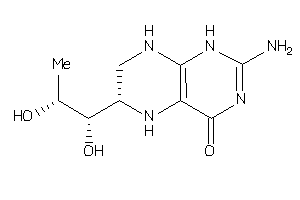
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-5.25 |
-21.45 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-3.31 |
-10.37 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.44 |
-11.69 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
G6PD-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.36 |
Binding ≤ 10μM
|
|
Q9GRG7-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.76 |
-6.06 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1367 |
0.39 |
Binding ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
3380 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1367 |
0.39 |
Binding ≤ 10μM
|
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z50573 |
Z50573
|
Xenopus Laevis |
3380 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.9 |
-6.34 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.65 |
-4.34 |
-17.19 |
6 |
8 |
0 |
137 |
239.235 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.07 |
-4.43 |
-43.98 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.07 |
-4.17 |
-38.96 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.65 |
-3.93 |
-16.78 |
6 |
8 |
0 |
137 |
239.235 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.65 |
-3.09 |
-42 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.65 |
-3.57 |
-39.05 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.65 |
-3.89 |
-16.51 |
6 |
8 |
0 |
137 |
239.235 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.65 |
-4.31 |
-17.21 |
6 |
8 |
0 |
137 |
239.235 |
2 |
↓
|
|
|
|
Analogs
-
3201876
-
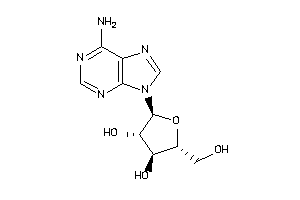
-
3201878
-
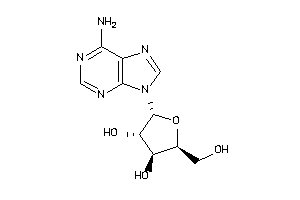
-
3830179
-
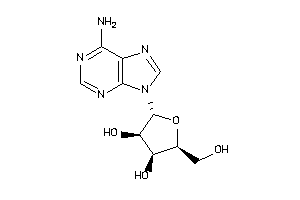
-
3978047
-
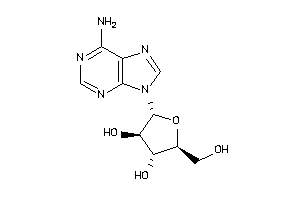
-
3978049
-
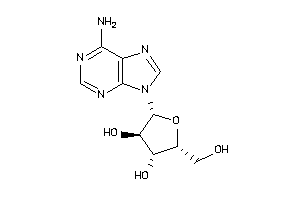
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
100 |
0.52 |
Binding ≤ 10μM
|
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8900 |
0.37 |
Binding ≤ 10μM
|
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.48 |
Functional ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.50 |
Functional ≤ 10μM
|
|
AA3R-1-E |
Adenosine A3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.48 |
Functional ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.50 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
6000 |
0.38 |
Functional ≤ 10μM
|
|
Z50588-2-O |
Canis Familiaris (cluster #2 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
700 |
0.45 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
10 |
0.59 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
30 |
0.55 |
Binding ≤ 1μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
37 |
0.55 |
Binding ≤ 1μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
100 |
0.52 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
10 |
0.59 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
30 |
0.55 |
Binding ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
37 |
0.55 |
Binding ≤ 10μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
100 |
0.52 |
Binding ≤ 10μM
|
|
ADK_TOXGO |
Q9TVW2
|
Adenosine Kinase, Toxgo |
8900 |
0.37 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
310 |
0.48 |
Functional ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
150 |
0.50 |
Functional ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
700 |
0.45 |
Functional ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
1040 |
0.44 |
Functional ≤ 10μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
150 |
0.50 |
Functional ≤ 10μM
|
|
Z50588 |
Z50588
|
Canis Familiaris |
0.12 |
0.73 |
Functional ≤ 10μM
|
|
Z80945 |
Z80945
|
HEp-2 (Laryngeal Carcinoma Cells) |
700 |
0.45 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
5940 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-4.39 |
-15.03 |
5 |
9 |
0 |
140 |
267.245 |
2 |
↓
|
|
|
|
Analogs
-
120294
-
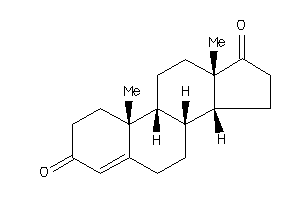
-
2046798
-
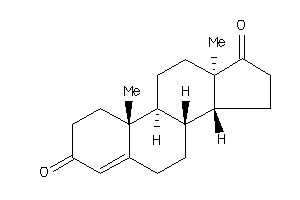
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.29 |
-10.68 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.06 |
9.39 |
-9.03 |
0 |
2 |
0 |
34 |
286.415 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.62 |
-3.47 |
-14.16 |
3 |
5 |
0 |
94 |
362.466 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.62 |
12.78 |
-1.53 |
1 |
1 |
0 |
20 |
386.664 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
5187769
-
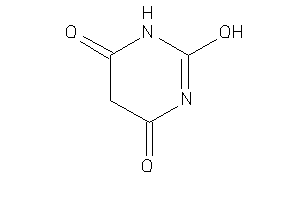
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
-2.01 |
-10.67 |
2 |
4 |
0 |
58 |
114.104 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
490588
-
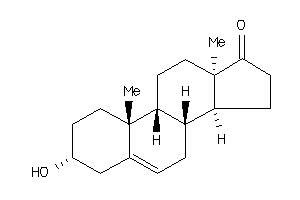
-
1036928
-
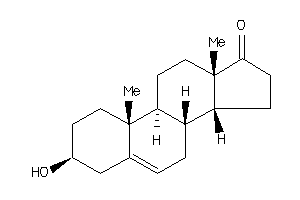
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.09 |
-6.2 |
1 |
2 |
0 |
37 |
288.431 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
6481415
-
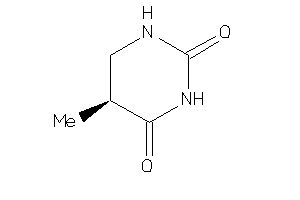
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.32 |
-1.43 |
-11.07 |
2 |
4 |
0 |
58 |
128.131 |
0 |
↓
|
|
|
|
Analogs
-
6437922
-
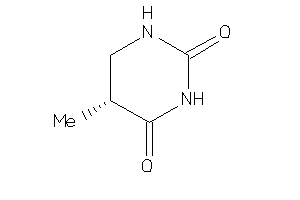
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.32 |
-1.46 |
-10.8 |
2 |
4 |
0 |
58 |
128.131 |
0 |
↓
|
|
|
|
Analogs
-
3983944
-
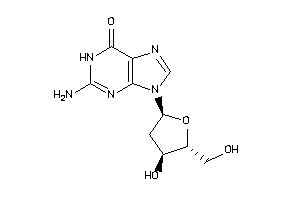
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.06 |
-2.94 |
-20.71 |
5 |
9 |
0 |
139 |
267.245 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
1532640
-
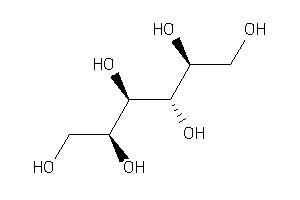
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.09 |
-12.44 |
-11.19 |
6 |
6 |
0 |
121 |
182.172 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-10.7 |
-6.85 |
5 |
6 |
0 |
110 |
180.156 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-10.59 |
-8.3 |
5 |
6 |
0 |
110 |
180.156 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
-6.46 |
-8.71 |
3 |
3 |
0 |
61 |
92.094 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
7.14 |
-7.63 |
1 |
2 |
0 |
37 |
270.372 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.53 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
|
SHBG-1-E |
Testis-specific Androgen-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Functional ≤ 10μM |
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Functional ≤ 10μM |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Functional ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z80226-1-O |
MCF7-2A (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 1μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 1μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 1μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 1μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 10μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 10μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.14 |
0.69 |
Functional ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
5.9 |
0.58 |
Functional ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
4.6 |
0.58 |
Functional ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
0.3 |
0.67 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
0.59 |
0.65 |
Functional ≤ 10μM
|
|
Z81068 |
Z81068
|
Ishikawa (Uterine Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80226 |
Z80226
|
MCF7-2A |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ADO_RAT |
Q9Z0U5
|
Aldehyde Oxidase, Rat |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
ADO_HUMAN |
Q06278
|
Aldehyde Oxidase, Human |
80 |
0.50 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
4.8 |
-5.19 |
2 |
2 |
0 |
40 |
272.388 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
1529206
-
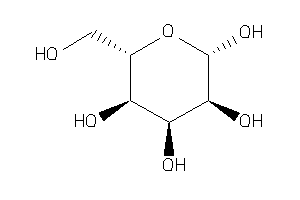
-
1532520
-
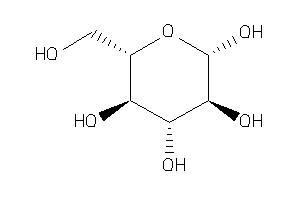
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-10.26 |
-8.6 |
5 |
6 |
0 |
110 |
180.156 |
1 |
↓
|
|
|
|
Analogs
-
1529206
-

-
1532520
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-10.58 |
-8.84 |
5 |
6 |
0 |
110 |
180.156 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
-9.07 |
-7.62 |
4 |
5 |
0 |
90 |
164.157 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
3830982
-
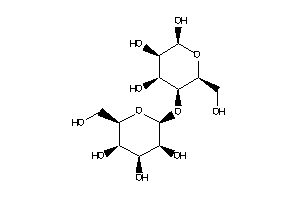
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.43 |
-19.4 |
-12.93 |
8 |
11 |
0 |
189 |
342.297 |
4 |
↓
|
|
|
|
Analogs
-
4015531
-
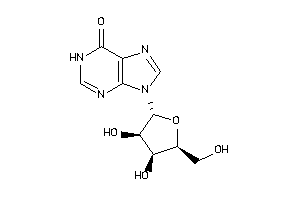
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.82 |
-4.43 |
-23.87 |
4 |
9 |
0 |
133 |
268.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.36 |
-6.52 |
-59.54 |
3 |
9 |
-1 |
137 |
267.221 |
2 |
↓
|
|