|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAKB1-1-E |
AMP-activated Protein Kinase, Beta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7700 |
0.42 |
Binding ≤ 10μM
|
|
AAKG1-1-E |
AMP-activated Protein Kinase, Gamma-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7700 |
0.42 |
Binding ≤ 10μM
|
|
AAPK2-1-E |
AMP-activated Protein Kinase, Alpha-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7700 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
4.84 |
-8.95 |
1 |
4 |
0 |
50 |
290.12 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
10.7 |
-14.87 |
0 |
2 |
0 |
6 |
320.574 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM6-2-E |
Metabotropic Glutamate Receptor 6 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.33 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
4500 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
0.24 |
-9.12 |
2 |
3 |
0 |
52 |
323.417 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KPCA_HUMAN |
P17252
|
Protein Kinase C Alpha, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB_HUMAN |
P05771
|
Protein Kinase C Beta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE_HUMAN |
Q02156
|
Protein Kinase C Epsilon, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG_HUMAN |
P05129
|
Protein Kinase C Gamma, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI_HUMAN |
P41743
|
Protein Kinase C Iota, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1_HUMAN |
Q15139
|
Protein Kinase C Mu, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3_HUMAN |
O94806
|
Protein Kinase C Nu, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT_HUMAN |
Q04759
|
Protein Kinase C Theta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ_HUMAN |
Q05513
|
Protein Kinase C Zeta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCA-2-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCB-2-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Binding ≤ 10μM
|
|
KGP1-1-E |
CGMP-dependent Protein Kinase 1 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KGP2-1-E |
CGMP-dependent Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCA-6-E |
Protein Kinase C Alpha (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB-3-E |
Protein Kinase C Beta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-4-E |
Protein Kinase C Delta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1-2-E |
Protein Kinase C Mu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3-2-E |
Protein Kinase C Nu (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE-4-E |
Protein Kinase C Epsilon (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG-2-E |
Protein Kinase C Gamma (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI-2-E |
Protein Kinase C Iota (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL-3-E |
Protein Kinase C Eta (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ-4-E |
Protein Kinase C Zeta (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
Q95J97-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5400 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
Q95J97_RABIT |
Q95J97
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Rabit |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
3000 |
0.39 |
Binding ≤ 10μM
|
|
KGP1_HUMAN |
Q13976
|
CGMP-dependent Protein Kinase 1 Beta, Human |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KGP2_HUMAN |
Q13237
|
CGMP-dependent Protein Kinase 2, Human |
5800 |
0.37 |
Binding ≤ 10μM
|
|
KPCA_HUMAN |
P17252
|
Protein Kinase C Alpha, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCB_HUMAN |
P05771
|
Protein Kinase C Beta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCE_HUMAN |
Q02156
|
Protein Kinase C Epsilon, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCG_HUMAN |
P05129
|
Protein Kinase C Gamma, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCI_HUMAN |
P41743
|
Protein Kinase C Iota, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD1_HUMAN |
Q15139
|
Protein Kinase C Mu, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCD3_HUMAN |
O94806
|
Protein Kinase C Nu, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCT_HUMAN |
Q04759
|
Protein Kinase C Theta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
KPCZ_HUMAN |
Q05513
|
Protein Kinase C Zeta, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5400 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ERCC5-1-E |
DNA Excision Repair Protein ERCC-5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
FEN1-1-E |
Flap Endonuclease 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
RNH1-2-E |
Ribonuclease H1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
2.48 |
-12.16 |
2 |
5 |
0 |
75 |
178.147 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RNH1-1-E |
Ribonuclease H1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
3.94 |
-12 |
1 |
4 |
0 |
58 |
177.159 |
0 |
↓
|
|
|
|
Analogs
-
4098705
-
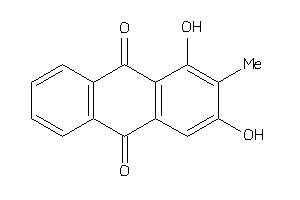
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
1.74 |
-10.81 |
3 |
5 |
0 |
95 |
270.24 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.01 |
2.52 |
-45.75 |
2 |
5 |
-1 |
98 |
269.232 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CSK21-3-E |
Casein Kinase II Alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Binding ≤ 10μM
|
|
CSK22-2-E |
Casein Kinase II Alpha (prime) (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Binding ≤ 10μM
|
|
CSK2B-2-E |
Casein Kinase II Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.43 |
6.37 |
-7.11 |
1 |
2 |
0 |
29 |
433.723 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
1.78 |
-4.16 |
1 |
1 |
0 |
20 |
130.093 |
0 |
↓
|
|
|
|
Analogs
-
4546563
-
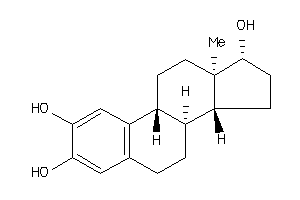
-
25757123
-
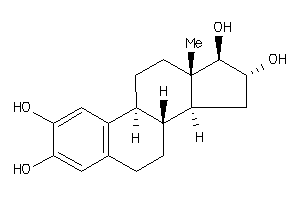
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
2.72 |
-7.57 |
3 |
3 |
0 |
61 |
288.387 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
-1.32 |
-9.83 |
2 |
3 |
0 |
58 |
214.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
6.05 |
-9.03 |
0 |
4 |
0 |
45 |
256.257 |
3 |
↓
|
|
|
|
Analogs
-
6117135
-
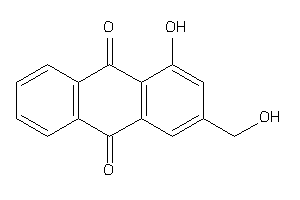
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AL1A7-1-E |
Aldehyde Dehydrogenase, Cytosolic 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM |
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
8700 |
0.35 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
10000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
1.23 |
-12.42 |
3 |
5 |
0 |
95 |
270.24 |
1 |
↓
|
|
|
|
|
|
|
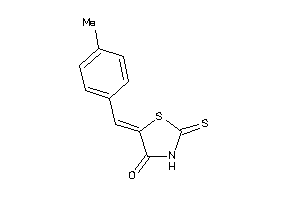
Analogs
-
1304776
-
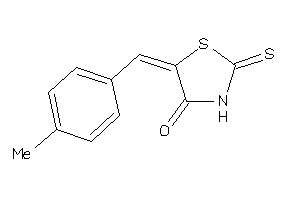
-
12407774
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
5.13 |
-8.07 |
1 |
2 |
0 |
33 |
221.306 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-3-E |
Hormone Sensitive Lipase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
370 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
5.26 |
-9.19 |
1 |
3 |
0 |
38 |
225.313 |
5 |
↓
|
|
|
|
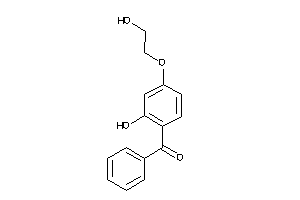
Analogs
-
5849525
-
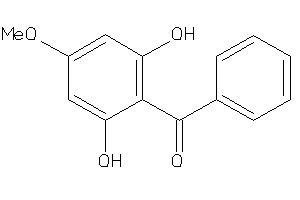
-
187
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS-2-E |
Hormone Sensitive Lipase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3250 |
0.45 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LIPS_RAT |
P15304
|
Hormone-sensitive Lipase, Rat |
3250 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
4.46 |
-8.1 |
1 |
3 |
0 |
47 |
228.247 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
5.24 |
-49.88 |
0 |
3 |
-1 |
49 |
227.239 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CTRA-2-E |
Alpha-chymotrypsin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.75 |
Binding ≤ 10μM
|
|
LIPS-2-E |
Hormone Sensitive Lipase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.52 |
Binding ≤ 10μM
|
|
MDHC-2-E |
Malate Dehydrogenase Cytoplasmic (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.67 |
Binding ≤ 10μM
|
|
MDHM-2-E |
Malate Dehydrogenase, Mitochondrial (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
9.11 |
-6.44 |
0 |
2 |
0 |
26 |
212.248 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q54A96-1-E |
Dihydroorotate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.41 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
-1.41 |
-12.99 |
2 |
6 |
0 |
75 |
291.314 |
2 |
↓
|
|
|
|
Analogs
-
13523524
-
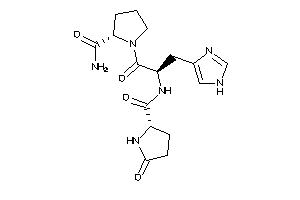
-
13523529
-
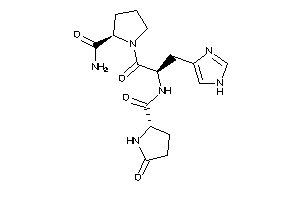
-
13523533
-
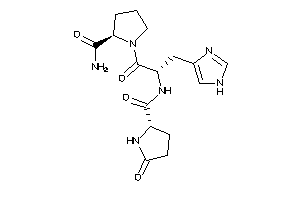
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.40 |
Binding ≤ 10μM
|
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 1μM
|
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 1μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 1μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 1μM
|
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
3 |
0.46 |
Functional ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
3 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.03 |
0.18 |
-23.59 |
5 |
10 |
0 |
150 |
362.39 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.03 |
0.01 |
-48.98 |
6 |
10 |
1 |
152 |
363.398 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.91 |
-8.71 |
-54.13 |
6 |
10 |
1 |
160 |
363.398 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
1.77 |
-3.35 |
1 |
1 |
0 |
20 |
130.093 |
0 |
↓
|
|
|
|
Analogs
-
37138061
-
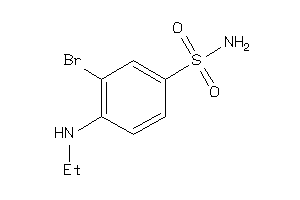
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
9650 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
73 |
0.77 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
175 |
0.73 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
68 |
0.77 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
-1.25 |
-8.6 |
4 |
4 |
0 |
86 |
330.001 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
UD2B7-1-E |
UDP-glucuronosyltransferase 2B7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
5.16 |
-1.81 |
1 |
1 |
0 |
20 |
222.372 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q54A96-1-E |
Dihydroorotate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
0.35 |
-9.19 |
1 |
2 |
0 |
29 |
340.22 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1367 |
0.39 |
Binding ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
3380 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
410 |
0.43 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1367 |
0.39 |
Binding ≤ 10μM
|
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z50573 |
Z50573
|
Xenopus Laevis |
3380 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.9 |
-6.34 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB3-3-E |
Estradiol 17-beta-dehydrogenase 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.43 |
Binding ≤ 10μM
|
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3760 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB3_HUMAN |
P37058
|
Estradiol 17-beta-dehydrogenase 3, Human |
182 |
0.45 |
Binding ≤ 1μM
|
|
DHB3_HUMAN |
P37058
|
Estradiol 17-beta-dehydrogenase 3, Human |
182 |
0.45 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
3760 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.62 |
-6.45 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
5765155
-
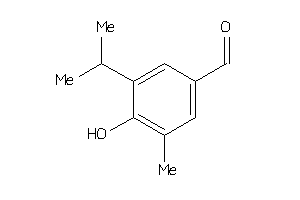
-
39233971
-
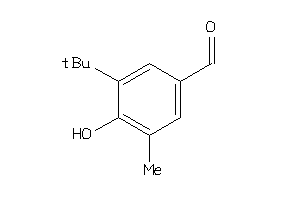
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
5800 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
5800 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
1.51 |
-6.94 |
1 |
2 |
0 |
37 |
206.285 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
1200 |
0.59 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1200 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
0.25 |
-2.45 |
1 |
1 |
0 |
20 |
304.171 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
0.77 |
-15.07 |
3 |
7 |
0 |
113 |
314.249 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
1.63 |
-43.84 |
2 |
7 |
-1 |
116 |
313.241 |
1 |
↓
|
|
|
|
Analogs
-
597316
-
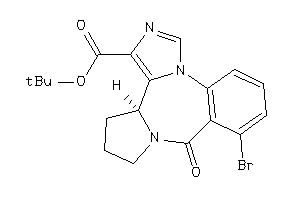
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
6 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
0.41 |
-15.67 |
0 |
6 |
0 |
64 |
418.291 |
3 |
↓
|
|
|
|
Analogs
-
11616881
-
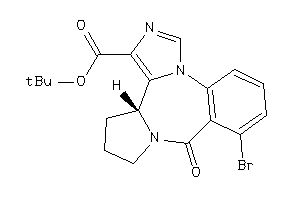
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
0.35 |
0.51 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
0.64 |
0.50 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
0.5 |
0.50 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
12.7 |
0.43 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
0.5 |
0.50 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
0.35 |
0.51 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
0.64 |
0.50 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
0.5 |
0.50 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
12.7 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
0.2 |
0.52 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
0.5 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
10.5 |
-15.44 |
0 |
6 |
0 |
64 |
418.291 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
180 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
8.96 |
-9.99 |
0 |
2 |
0 |
30 |
236.27 |
1 |
↓
|
|
|
|
Analogs
-
1961543
-
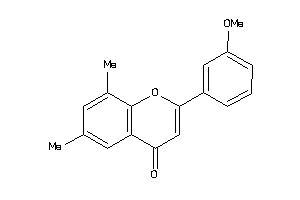
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
99 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
2.42 |
-10.66 |
0 |
3 |
0 |
39 |
266.296 |
2 |
↓
|
|
|
|
Analogs
-
39119523
-
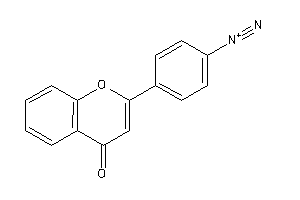
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
2600 |
0.41 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
2600 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
6.84 |
-11.4 |
2 |
3 |
0 |
56 |
251.285 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
820 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
2.64 |
-11.94 |
0 |
3 |
0 |
39 |
266.296 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
390 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
6.1 |
-11.06 |
1 |
3 |
0 |
50 |
252.269 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
12 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
1.65 |
-10.08 |
0 |
2 |
0 |
30 |
315.166 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
1200 |
0.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1200 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
6.83 |
-10.39 |
2 |
3 |
0 |
56 |
251.285 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
4400 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
4400 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
2.73 |
-9.87 |
0 |
2 |
0 |
30 |
250.297 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.49 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
574 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-4-E |
GABA Receptor Alpha-1 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.58 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5 |
0.58 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
12 |
0.55 |
Binding ≤ 1μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
531 |
0.44 |
Binding ≤ 1μM
|
|
GBRA1_BOVIN |
P08219
|
GABA Receptor Alpha-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
15 |
0.55 |
Binding ≤ 1μM
|
|
GBRA2_BOVIN |
P10063
|
GABA Receptor Alpha-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA3_BOVIN |
P10064
|
GABA Receptor Alpha-3 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
13.9 |
0.55 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA4_BOVIN |
P20237
|
GABA Receptor Alpha-4 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
23 |
0.53 |
Binding ≤ 1μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
2.1 |
0.61 |
Binding ≤ 1μM
|
|
GBRB1_BOVIN |
P08220
|
GABA Receptor Beta-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRG2_BOVIN |
P22300
|
GABA Receptor Gamma-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10.5 |
0.56 |
Binding ≤ 1μM
|
|
TSPO_RAT |
P16257
|
Peripheral-type Benzodiazepine Receptor, Rat |
213 |
0.47 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA1_BOVIN |
P08219
|
GABA Receptor Alpha-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_BOVIN |
P10063
|
GABA Receptor Alpha-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
15 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_BOVIN |
P10064
|
GABA Receptor Alpha-3 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
13.9 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA4_BOVIN |
P20237
|
GABA Receptor Alpha-4 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
23 |
0.53 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
2.1 |
0.61 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRB1_BOVIN |
P08220
|
GABA Receptor Beta-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRG2_BOVIN |
P22300
|
GABA Receptor Gamma-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10.5 |
0.56 |
Binding ≤ 10μM
|
|
TSPO_RAT |
P16257
|
Peripheral-type Benzodiazepine Receptor, Rat |
213 |
0.47 |
Binding ≤ 10μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
5 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
6.91 |
-8.26 |
0 |
3 |
0 |
33 |
284.746 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.74 |
7.78 |
-30.37 |
1 |
3 |
1 |
34 |
285.754 |
1 |
↓
|
|
|
|
Analogs
-
34461098
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAN2-1-E |
Calpain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
CANX-1-E |
Calpain 1/2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
0.79 |
-13.07 |
1 |
4 |
0 |
55 |
283.327 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.16 |
-15.04 |
3 |
7 |
0 |
109 |
330.292 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.53 |
1.45 |
-45.8 |
2 |
7 |
-1 |
112 |
329.284 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.39 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.39 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
4860 |
0.32 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
83 |
0.43 |
Functional ≤ 10μM
|
|
Z80125-9-O |
DU-145 (Prostate Carcinoma) (cluster #9 Of 9), Other |
Other |
34 |
0.45 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
31 |
0.46 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
560 |
0.38 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
41 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
437 |
0.39 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
394 |
0.39 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
170 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
300 |
0.40 |
Functional ≤ 10μM
|
|
Z80798-1-O |
CPT30 Cell Line (cluster #1 Of 1), Other |
Other |
37 |
0.45 |
Functional ≤ 10μM
|
|
Z80900-1-O |
HA22T Cell Line (cluster #1 Of 1), Other |
Other |
3706 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
33 |
0.46 |
Functional ≤ 10μM
|
|
Z80967-2-O |
Hone-1 Cell Line (cluster #2 Of 2), Other |
Other |
87 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
35 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
30 |
0.46 |
Functional ≤ 10μM
|
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
28 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
4.04 |
-11.69 |
1 |
6 |
0 |
74 |
318.325 |
6 |
↓
|
|
|
|
Analogs
-
6483405
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.30 |
Binding ≤ 10μM |
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
1.66 |
-15.02 |
3 |
8 |
0 |
119 |
360.318 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
1.83 |
1.54 |
-25.17 |
3 |
8 |
0 |
119 |
360.318 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
2.92 |
-62.02 |
2 |
8 |
-1 |
121 |
359.31 |
4 |
↓
|
|
|
|
Analogs
-
34527023
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.30 |
Functional ≤ 10μM
|
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.30 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.30 |
Functional ≤ 10μM
|
|
TBB2B-2-E |
Tubulin Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.30 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
1111 |
0.32 |
Functional ≤ 10μM |
|
Z80125-4-O |
DU-145 (Prostate Carcinoma) (cluster #4 Of 9), Other |
Other |
839 |
0.33 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
340 |
0.35 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
190 |
0.36 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
786 |
0.33 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
960 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
350 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
170 |
0.36 |
Functional ≤ 10μM |
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
160 |
0.37 |
Functional ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
658 |
0.33 |
Functional ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
270 |
0.35 |
Functional ≤ 10μM |
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
190 |
0.36 |
Functional ≤ 10μM |
|
Z81335-4-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #4 Of 5), Other |
Other |
340 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
3.28 |
-12.76 |
3 |
7 |
0 |
101 |
371.418 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.33 |
3.22 |
-44.02 |
2 |
7 |
-1 |
103 |
370.41 |
6 |
↓
|
|