|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.17 |
5.79 |
-29.19 |
3 |
4 |
0 |
73 |
218.256 |
4 |
↓
|
|
|
|
Analogs
-
3881796
-
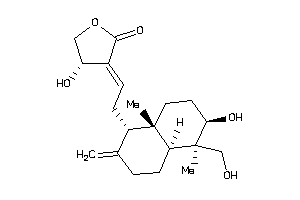
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.5 |
-12.88 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
2320 |
0.36 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8910 |
0.32 |
Binding ≤ 10μM
|
|
CAH2-15-E |
Carbonic Anhydrase II (cluster #15 Of 15), Eukaryotic |
Eukaryotes |
2180 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
9080 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
7590 |
0.33 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
7060 |
0.33 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
6320 |
0.33 |
Binding ≤ 10μM
|
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
9370 |
0.32 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
ERG1-3-E |
Squalene Monooxygenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
FGR-2-E |
Tyrosine-protein Kinase FGR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.32 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
7500 |
0.33 |
Binding ≤ 10μM
|
|
KAPCA-3-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KAPCB-3-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KAPCG-2-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KSYK-2-E |
Tyrosine-protein Kinase SYK (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.35 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.42 |
Binding ≤ 10μM
|
|
Z50136-3-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #3 Of 3), Other |
Other |
300 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
330 |
0.41 |
Functional ≤ 10μM
|
|
Z50426-4-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #4 Of 9), Other |
Other |
970 |
0.38 |
Functional ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
5100 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
40 |
0.47 |
Binding ≤ 1μM
|
|
CSK21_RAT |
P19139
|
Casein Kinase II Alpha, Rat |
20 |
0.49 |
Binding ≤ 1μM
|
|
CSK2B_RAT |
P67874
|
Casein Kinase II Beta, Rat |
20 |
0.49 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
300 |
0.42 |
Binding ≤ 1μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2320 |
0.36 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
2180 |
0.36 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
9080 |
0.32 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
9370 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
7590 |
0.33 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
7060 |
0.33 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6320 |
0.33 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
8910 |
0.32 |
Binding ≤ 10μM
|
|
CSK21_RAT |
P19139
|
Casein Kinase II Alpha, Rat |
20 |
0.49 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
40 |
0.47 |
Binding ≤ 10μM
|
|
CSK2B_RAT |
P67874
|
Casein Kinase II Beta, Rat |
20 |
0.49 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
7500 |
0.33 |
Binding ≤ 10μM
|
|
Q7ZJM1_9HIV1 |
Q7ZJM1
|
Human Immunodeficiency Virus Type 1 Integrase, 9hiv1 |
5100 |
0.34 |
Binding ≤ 10μM
|
|
ERG1_RAT |
P52020
|
Squalene Monooxygenase, Rat |
2000 |
0.36 |
Binding ≤ 10μM
|
|
FGR_RAT |
Q6P6U0
|
Tyrosine-protein Kinase FGR, Rat |
9400 |
0.32 |
Binding ≤ 10μM
|
|
LYN_RAT |
Q07014
|
Tyrosine-protein Kinase Lyn, Rat |
2900 |
0.35 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
300 |
0.42 |
Binding ≤ 10μM
|
|
KSYK_RAT |
Q64725
|
Tyrosine-protein Kinase SYK, Rat |
4300 |
0.34 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
180 |
0.43 |
Functional ≤ 10μM
|
|
Z50136 |
Z50136
|
Plasmodium Falciparum (isolate FcB1 / Columbia) |
300 |
0.42 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
970 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-1.09 |
-13.29 |
4 |
8 |
0 |
141 |
302.194 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.49 |
-0.46 |
-37.99 |
3 |
8 |
-1 |
144 |
301.186 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.94 |
-0.07 |
-45.06 |
3 |
8 |
-1 |
144 |
301.186 |
0 |
↓
|
|
|
|
Analogs
-
3872686
-
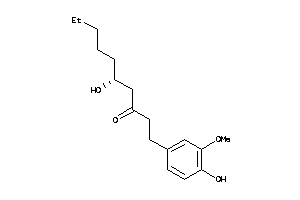
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
6.04 |
-12.03 |
2 |
4 |
0 |
67 |
294.391 |
10 |
↓
|
|
|
|
Analogs
-
1531846
-
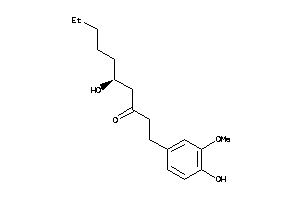
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
6.04 |
-12.05 |
2 |
4 |
0 |
67 |
294.391 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
12 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
3.61 |
-6.89 |
1 |
3 |
0 |
37 |
214.268 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.51 |
4.94 |
-100.78 |
3 |
3 |
2 |
40 |
216.284 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
1.46 |
-7.2 |
2 |
3 |
0 |
48 |
200.241 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
1.77 |
3.41 |
-48.19 |
2 |
3 |
0 |
45 |
200.241 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
1.97 |
2.48 |
-31.67 |
3 |
3 |
1 |
46 |
201.249 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
2.68 |
-7.61 |
2 |
3 |
0 |
49 |
198.225 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
2.09 |
3.1 |
-33.34 |
3 |
3 |
1 |
50 |
199.233 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.09 |
3.44 |
-44.12 |
1 |
3 |
-1 |
52 |
197.217 |
0 |
↓
|
|
|
|
Analogs
-
5195818
-
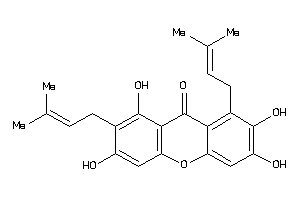
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.32 |
6.65 |
-10.92 |
3 |
6 |
0 |
100 |
410.466 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.42 |
7.43 |
-44.09 |
2 |
6 |
-1 |
103 |
409.458 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYRO-7-F |
Tyrosinase (cluster #7 Of 8), Fungal |
Fungi |
3680 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.16 |
1.04 |
-45.07 |
4 |
6 |
0 |
110 |
198.178 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.00 |
8.44 |
-11.93 |
0 |
7 |
0 |
77 |
425.436 |
7 |
↓
|
|
|
|
Analogs
-
4245611
-
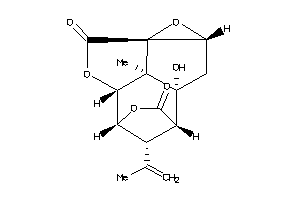
-
17027441
-
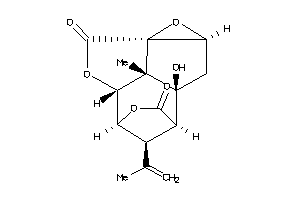
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA2-1-E |
Glycine Receptor Subunit Alpha-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA2_RAT |
P22771
|
Glycine Receptor Alpha-2 Chain, Rat |
2400 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.21 |
2.83 |
-16.67 |
1 |
6 |
0 |
85 |
292.287 |
1 |
↓
|
|
|
|
Analogs
-
34583707
-
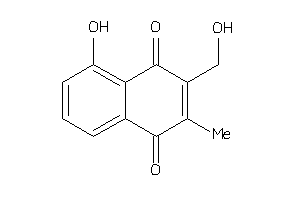
-
39283830
-
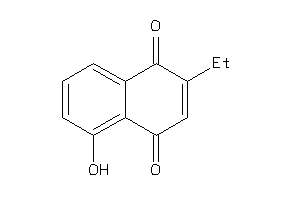
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
270 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
4.24 |
-7.85 |
1 |
3 |
0 |
54 |
188.182 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
7.13 |
-12.93 |
0 |
4 |
0 |
63 |
293.298 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.16 |
3.8 |
-22.24 |
0 |
4 |
0 |
57 |
294.306 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.36 |
9.56 |
-59.86 |
0 |
4 |
-1 |
63 |
293.298 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.16 |
1.03 |
-42.84 |
4 |
6 |
0 |
110 |
198.178 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
3881796
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.68 |
-13.02 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
-
3881796
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
-3.14 |
-14.34 |
3 |
5 |
0 |
86 |
350.455 |
3 |
↓
|
|
|
|
Analogs
-
3881796
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
-3.79 |
-14.69 |
3 |
5 |
0 |
86 |
350.455 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4482687
-
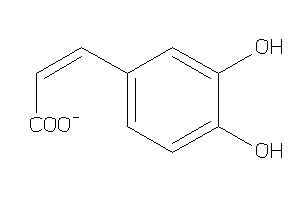
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8710 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
7870 |
0.55 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM
|
|
O49150-1-E |
5-lipoxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3060 |
0.59 |
Binding ≤ 10μM
|
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
2900 |
0.60 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.72 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O49150_SOLTU |
O49150
|
5-lipoxygenase, Soltu |
3000 |
0.59 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
7870 |
0.55 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
8710 |
0.54 |
Binding ≤ 10μM
|
|
PTN1_HUMAN |
P18031
|
Protein-tyrosine Phosphatase 1B, Human |
3060 |
0.59 |
Binding ≤ 10μM
|
|
LOX1_SOYBN |
P08170
|
Seed Lipoxygenase-1, Soybn |
3000 |
0.59 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
190 |
0.72 |
Functional ≤ 10μM
|
|
Z100766 |
Z100766
|
Radical Scavenging Activity |
2800 |
0.60 |
Functional ≤ 10μM
|
|
Z50185 |
Z50185
|
Staphylococcus Aureus |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186 |
Z50186
|
Staphylococcus Epidermidis |
2780 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.43 |
-49.59 |
2 |
4 |
-1 |
81 |
179.151 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
33799539
-
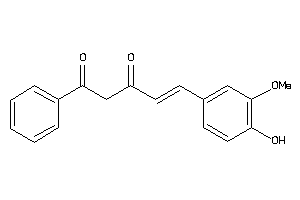
-
44608728
-
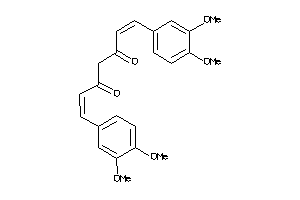
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTSI-1-B |
CpG DNA Methylase (cluster #1 Of 2), Bacterial |
Bacteria |
30 |
0.39 |
Binding ≤ 10μM
|
|
A4-1-E |
Beta Amyloid A4 Protein (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
800 |
0.32 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2410 |
0.29 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
3480 |
0.28 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6850 |
0.27 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5090 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
380 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4970 |
0.28 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9460 |
0.26 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
9940 |
0.26 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
9300 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4050 |
0.28 |
Binding ≤ 10μM
|
|
DHB3-1-E |
Estradiol 17-beta-dehydrogenase 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
KPCE-5-E |
Protein Kinase C Epsilon (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
8810 |
0.26 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8000 |
0.26 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.26 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
PGH1-6-E |
Cyclooxygenase-1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8800 |
0.26 |
Binding ≤ 10μM
|
|
Q8HY88-1-E |
Potassium Channel Subfamily K Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
930 |
0.31 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4300 |
0.28 |
ADME/T ≤ 10μM
|
|
Z103192-1-O |
Trypanosoma Evansi (cluster #1 Of 2), Other |
Other |
2000 |
0.30 |
Functional ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
4800 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
4210 |
0.28 |
Functional ≤ 10μM
|
|
Z50515-1-O |
Human Herpesvirus 2 (cluster #1 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50518-1-O |
Human Herpesvirus 4 (cluster #1 Of 5), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50600-2-O |
Vaccinia Virus (cluster #2 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50602-1-O |
Human Herpesvirus 1 (cluster #1 Of 5), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z50651-2-O |
Vesicular Stomatitis Virus (cluster #2 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50658-4-O |
Human Immunodeficiency Virus 2 (cluster #4 Of 4), Other |
Other |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 11), Other |
Other |
6810 |
0.27 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5580 |
0.27 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
9700 |
0.26 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7700 |
0.27 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
8300 |
0.26 |
Functional ≤ 10μM
|
|
Z80548-3-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
1210 |
0.31 |
Functional ≤ 10μM |
|
Z80612-1-O |
2008 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.27 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
7600 |
0.27 |
Functional ≤ 10μM
|
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
9000 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 4), Other |
Other |
8700 |
0.26 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A4_HUMAN |
P05067
|
Beta Amyloid A4 Protein, Human |
0.208 |
0.50 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
380 |
0.33 |
Binding ≤ 1μM
|
|
MTSI_SPISQ |
P15840
|
CpG DNA Methylase, Spisq |
30 |
0.39 |
Binding ≤ 1μM
|
|
Q8HY88_BOVIN |
Q8HY88
|
Potassium Channel Subfamily K Member 2, Bovin |
930 |
0.31 |
Binding ≤ 1μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
8000 |
0.26 |
Binding ≤ 10μM
|
|
A4_HUMAN |
P05067
|
Beta Amyloid A4 Protein, Human |
0.208 |
0.50 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
5090 |
0.27 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2410 |
0.29 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
380 |
0.33 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4970 |
0.28 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
4050 |
0.28 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9460 |
0.26 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
9940 |
0.26 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
9300 |
0.26 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3480 |
0.28 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
6850 |
0.27 |
Binding ≤ 10μM
|
|
MTSI_SPISQ |
P15840
|
CpG DNA Methylase, Spisq |
30 |
0.39 |
Binding ≤ 10μM
|
|
PGH1_HUMAN |
P23219
|
Cyclooxygenase-1, Human |
8800 |
0.26 |
Binding ≤ 10μM
|
|
LGUL_HUMAN |
Q04760
|
Glyoxalase I, Human |
10000 |
0.26 |
Binding ≤ 10μM
|
|
MMP9_HUMAN |
P14780
|
Matrix Metalloproteinase 9, Human |
8500 |
0.26 |
Binding ≤ 10μM
|
|
NOS2_HUMAN |
P35228
|
Nitric Oxide Synthase, Inducible, Human |
6000 |
0.27 |
Binding ≤ 10μM
|
|
Q8HY88_BOVIN |
Q8HY88
|
Potassium Channel Subfamily K Member 2, Bovin |
930 |
0.31 |
Binding ≤ 10μM
|
|
KPCE_HUMAN |
Q02156
|
Protein Kinase C Epsilon, Human |
8810 |
0.26 |
Binding ≤ 10μM
|
|
DHB3_RAT |
O54939
|
Testosterone 17-beta-dehydrogenase 3, Rat |
2300 |
0.29 |
Binding ≤ 10μM
|
|
Z80612 |
Z80612
|
2008 (Ovarian Carcinoma Cells) |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z50602 |
Z50602
|
Human Herpesvirus 1 |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50515 |
Z50515
|
Human Herpesvirus 2 |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50518 |
Z50518
|
Human Herpesvirus 4 |
10.2 |
0.41 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z50658 |
Z50658
|
Human Immunodeficiency Virus 2 |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
6810 |
0.27 |
Functional ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
3800 |
0.28 |
Functional ≤ 10μM
|
|
Z81186 |
Z81186
|
LS174T (Colon Adencocarcinoma Cells) |
6500 |
0.27 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
5580 |
0.27 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
7600 |
0.27 |
Functional ≤ 10μM
|
|
Z80244 |
Z80244
|
MDA-MB-468 (Breast Adenocarcinoma) |
9700 |
0.26 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
7700 |
0.27 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3250 |
0.28 |
Functional ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
8300 |
0.26 |
Functional ≤ 10μM
|
|
Z80548 |
Z80548
|
THP-1 (Acute Monocytic Leukemia Cells) |
1210 |
0.31 |
Functional ≤ 10μM
|
|
Z50420 |
Z50420
|
Trypanosoma Brucei Brucei |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z103192 |
Z103192
|
Trypanosoma Evansi |
2000 |
0.30 |
Functional ≤ 10μM
|
|
Z50600 |
Z50600
|
Vaccinia Virus |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50651 |
Z50651
|
Vesicular Stomatitis Virus |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
8700 |
0.26 |
ADME/T ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
4300 |
0.28 |
ADME/T ≤ 10μM
|
|
Z80193 |
Z80193
|
L1210 (Lymphocytic Leukemia Cells) |
9000 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
7.14 |
-22.28 |
2 |
6 |
0 |
93 |
368.385 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
5.47 |
-58.07 |
2 |
6 |
-1 |
99 |
367.377 |
7 |
↓
|
|
|
|
Analogs
-
31970705
-
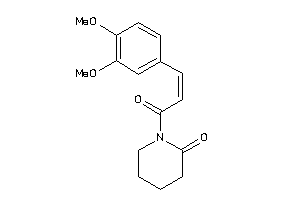
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
7.79 |
-21.88 |
0 |
6 |
0 |
65 |
319.357 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
9.87 |
-54.01 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
7.94 |
-7.27 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
288 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.55 |
12.56 |
-14.64 |
0 |
8 |
0 |
62 |
622.762 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.55 |
15.24 |
-57.28 |
1 |
8 |
1 |
63 |
623.77 |
4 |
↓
|
|
|
|
Analogs
-
8382399
-
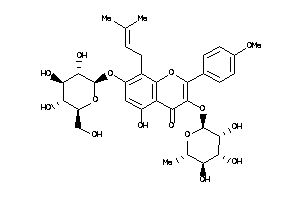
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
-15.05 |
-23.81 |
8 |
15 |
0 |
238 |
676.668 |
9 |
↓
|
|
|
|
Analogs
-
8382399
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-3.67 |
-21.04 |
8 |
15 |
0 |
238 |
676.668 |
9 |
↓
|
|
|
|
Analogs
-
8382399
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-3.52 |
-19.13 |
8 |
15 |
0 |
238 |
676.668 |
9 |
↓
|
|
|
|
Analogs
-
8382399
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-2.52 |
-21.18 |
8 |
15 |
0 |
238 |
676.668 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.89 |
-14.36 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.86 |
-13.22 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.48 |
-13.17 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.75 |
-13.54 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7790 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9340 |
0.44 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1480 |
0.51 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.46 |
Binding ≤ 10μM
|
|
ADA2A-4-E |
Alpha-2a Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2540 |
0.49 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1130 |
0.52 |
Binding ≤ 10μM
|
|
ADA2C-4-E |
Alpha-2c Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
810 |
0.53 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.73 |
Binding ≤ 10μM
|
|
AOFB-6-E |
Monoamine Oxidase B (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
49 |
0.64 |
Binding ≤ 10μM
|
|
DYR1A-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.62 |
Binding ≤ 10μM
|
|
DYRK2-1-E |
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.53 |
Binding ≤ 10μM
|
|
DYRK3-1-E |
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
NISCH-3-E |
Nischarin (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
49 |
0.64 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.47 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3260 |
0.48 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
50 |
0.64 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
520 |
0.55 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
810 |
0.53 |
Binding ≤ 1μM
|
|
DYR1A_HUMAN |
Q13627
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 1A, Human |
80 |
0.62 |
Binding ≤ 1μM
|
|
DYRK2_HUMAN |
Q92630
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 2, Human |
900 |
0.53 |
Binding ≤ 1μM
|
|
DYRK3_HUMAN |
O43781
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 3, Human |
800 |
0.53 |
Binding ≤ 1μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
16.9 |
0.68 |
Binding ≤ 1μM
|
|
AOFA_RAT |
P21396
|
Monoamine Oxidase A, Rat |
49 |
0.64 |
Binding ≤ 1μM
|
|
AOFB_RAT |
P19643
|
Monoamine Oxidase B, Rat |
49 |
0.64 |
Binding ≤ 1μM
|
|
NISCH_HUMAN |
Q9Y2I1
|
Nischarin, Human |
22 |
0.67 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
2540 |
0.49 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
1130 |
0.52 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
810 |
0.53 |
Binding ≤ 10μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
1500 |
0.51 |
Binding ≤ 10μM
|
|
DYR1A_HUMAN |
Q13627
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 1A, Human |
80 |
0.62 |
Binding ≤ 10μM
|
|
DYRK2_HUMAN |
Q92630
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 2, Human |
900 |
0.53 |
Binding ≤ 10μM
|
|
DYRK3_HUMAN |
O43781
|
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 3, Human |
800 |
0.53 |
Binding ≤ 10μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
16.9 |
0.68 |
Binding ≤ 10μM
|
|
AOFA_RAT |
P21396
|
Monoamine Oxidase A, Rat |
49 |
0.64 |
Binding ≤ 10μM
|
|
AOFB_RAT |
P19643
|
Monoamine Oxidase B, Rat |
49 |
0.64 |
Binding ≤ 10μM
|
|
NISCH_HUMAN |
Q9Y2I1
|
Nischarin, Human |
22 |
0.67 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
3260 |
0.48 |
Binding ≤ 10μM
|
|
PIM3_HUMAN |
Q86V86
|
Serine/threonine-protein Kinase PIM3, Human |
4300 |
0.47 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
7790 |
0.45 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
9340 |
0.44 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
1480 |
0.51 |
Binding ≤ 10μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
5500 |
0.46 |
Binding ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
520 |
0.55 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
182.4 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
5.25 |
-32.65 |
2 |
3 |
1 |
39 |
213.26 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.63 |
4.83 |
-7.3 |
1 |
3 |
0 |
38 |
212.252 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
13.24 |
-77.43 |
2 |
2 |
2 |
9 |
358.614 |
1 |
↓
|
|
|
|
Analogs
-
4349851
-
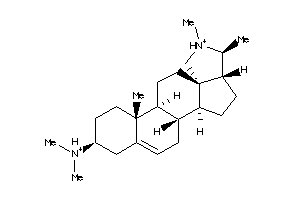
-
4577166
-
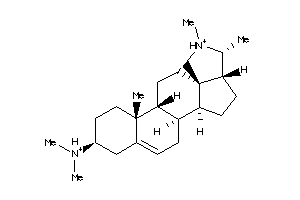
-
4577167
-
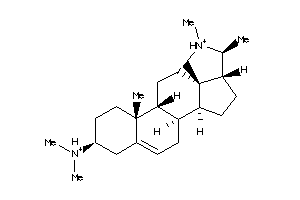
-
5529758
-
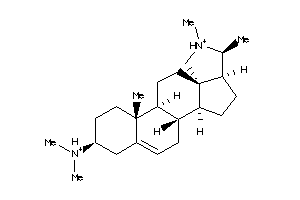
-
5752284
-
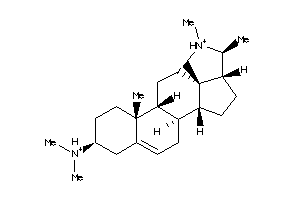
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
660.693448 |
0.33 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
10.4712855 |
0.43 |
Binding ≤ 1μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
3.4673685 |
0.46 |
Binding ≤ 1μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
12.3026877 |
0.43 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
660.693448 |
0.33 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
10.4712855 |
0.43 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
10000 |
0.27 |
Binding ≤ 10μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
10000 |
0.27 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
3.4673685 |
0.46 |
Binding ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
12.3026877 |
0.43 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
10000 |
0.27 |
Binding ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
1040 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
13.04 |
-76.57 |
2 |
2 |
2 |
9 |
358.614 |
1 |
↓
|
|
|
|
|