|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
5130 |
0.26 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5130 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.27 |
9.1 |
-12.31 |
3 |
4 |
0 |
78 |
392.495 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100751-1-O |
PMNL (Polymorphonuclear Leukocytes) (cluster #1 Of 2), Other |
Other |
6000 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100751 |
Z100751
|
PMNL (Polymorphonuclear Leukocytes) |
6000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
4.16 |
-17 |
3 |
8 |
0 |
130 |
374.345 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
5.73 |
-110.55 |
1 |
8 |
-2 |
136 |
372.329 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.55 |
4.95 |
-46.6 |
2 |
8 |
-1 |
133 |
373.337 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
800 |
0.33 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
2000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
0.41 |
-10.11 |
0 |
5 |
0 |
40 |
349.386 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
2.11 |
-11.85 |
0 |
6 |
0 |
57 |
405.45 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
3.62 |
-5.44 |
1 |
2 |
0 |
29 |
164.204 |
3 |
↓
|
|
|
|
Analogs
-
1529208
-
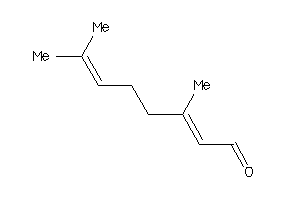
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
3.04 |
-5.54 |
0 |
1 |
0 |
17 |
152.237 |
4 |
↓
|
|
|
|
Analogs
-
12358789
-
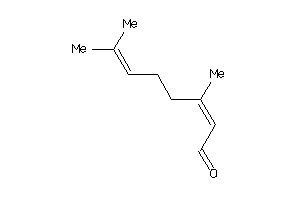
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
6.72 |
-6.07 |
0 |
1 |
0 |
17 |
152.237 |
4 |
↓
|
|
|
|
Analogs
-
1531533
-
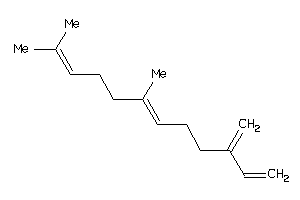
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
7.33 |
-0.89 |
0 |
0 |
0 |
0 |
136.238 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
5849312
-
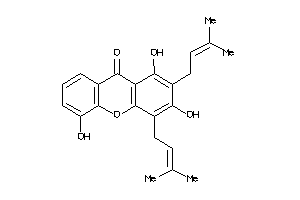
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
3600 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
6.45 |
-16.73 |
4 |
6 |
0 |
111 |
396.439 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.07 |
7.2 |
-72.36 |
3 |
6 |
-1 |
114 |
395.431 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.07 |
6.8 |
-48.13 |
3 |
6 |
-1 |
114 |
395.431 |
4 |
↓
|
|
|
|
Analogs
-
39289884
-
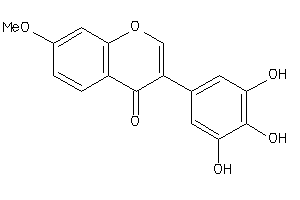
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
2.64 |
-12.54 |
2 |
5 |
0 |
80 |
284.267 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.64 |
2.74 |
-43.59 |
1 |
5 |
-1 |
83 |
283.259 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
6.13 |
-8.97 |
3 |
5 |
0 |
87 |
408.494 |
5 |
↓
|
|
|
|
Analogs
-
2149675
-
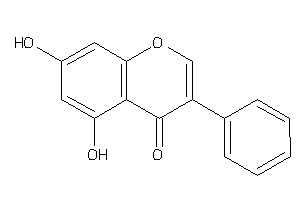
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.37 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.35 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
AOFB-4-E |
Monoamine Oxidase B (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2210 |
0.40 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.39 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
990 |
0.42 |
Binding ≤ 10μM
|
|
ESR2-4-E |
Estrogen Receptor Beta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
990 |
0.42 |
Binding ≤ 10μM
|
|
MGA-3-E |
Maltase-glucoamylase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2610 |
0.39 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.36 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
950 |
0.42 |
Functional ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
956 |
0.42 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.45 |
ADME/T ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
4200 |
0.38 |
Functional ≤ 10μM
|
|
Z80030-1-O |
ANN-1 (cluster #1 Of 2), Other |
Other |
8000 |
0.36 |
Functional ≤ 10μM
|
|
Z80037-1-O |
Balb/MK (cluster #1 Of 1), Other |
Other |
9100 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
510 |
0.44 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
956 |
0.42 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
1000 |
0.42 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
20 |
0.54 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1.3 |
0.62 |
Binding ≤ 1μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
395 |
0.45 |
Binding ≤ 1μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
900 |
0.42 |
Binding ≤ 1μM
|
|
AOFB_HUMAN |
P27338
|
Monoamine Oxidase B, Human |
900 |
0.42 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
5000 |
0.37 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
6900 |
0.36 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
1000 |
0.42 |
Binding ≤ 10μM
|
|
DHB1_HUMAN |
P14061
|
Estradiol 17-beta-dehydrogenase 1, Human |
2210 |
0.40 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
1145 |
0.42 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1.3 |
0.62 |
Binding ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
395 |
0.45 |
Binding ≤ 10μM
|
|
Q965D7_PLAFA |
Q965D7
|
Fatty Acid Synthase, Plafa |
7000 |
0.36 |
Binding ≤ 10μM
|
|
MGA_HUMAN |
O43451
|
Maltase-glucoamylase, Human |
2610 |
0.39 |
Binding ≤ 10μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
900 |
0.42 |
Binding ≤ 10μM
|
|
AOFB_HUMAN |
P27338
|
Monoamine Oxidase B, Human |
900 |
0.42 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
10000 |
0.35 |
Binding ≤ 10μM
|
|
Z80030 |
Z80030
|
ANN-1 |
8000 |
0.36 |
Functional ≤ 10μM
|
|
Z80037 |
Z80037
|
Balb/MK |
9100 |
0.35 |
Functional ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
810 |
0.43 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1.7 |
0.61 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
73 |
0.50 |
Functional ≤ 10μM
|
|
Z81068 |
Z81068
|
Ishikawa (Uterine Carcinoma Cells) |
510 |
0.44 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z50420 |
Z50420
|
Trypanosoma Brucei Brucei |
4200 |
0.38 |
Functional ≤ 10μM
|
|
ADO_HUMAN |
Q06278
|
Aldehyde Oxidase, Human |
340 |
0.45 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.08 |
-13.3 |
3 |
5 |
0 |
91 |
270.24 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
5.79 |
-13.65 |
2 |
5 |
0 |
80 |
336.343 |
1 |
↓
|
|
|
|
Analogs
-
39289597
-
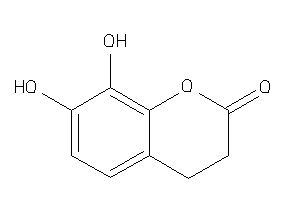
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
5.3 |
-9.92 |
0 |
3 |
0 |
36 |
178.187 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4720 |
0.36 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5030 |
0.35 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
450 |
0.42 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
5030 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4720 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
-4.94 |
-11.51 |
5 |
6 |
0 |
110 |
290.271 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
4096340
-
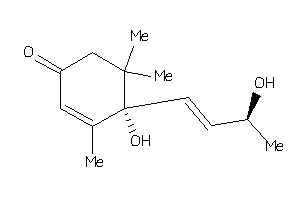
-
4095719
-
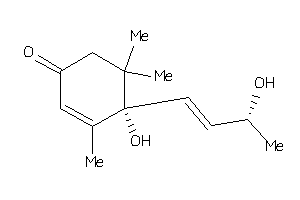
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
1.61 |
-7.75 |
2 |
3 |
0 |
58 |
224.3 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PA2GA-1-E |
Phospholipase A2, Membrane Associated (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.19 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PA2GA_HUMAN |
P14555
|
Phospholipase A2 Group IIA, Human |
3500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
4.08 |
-39.7 |
5 |
10 |
0 |
171 |
538.464 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.18 |
5.09 |
-85.16 |
4 |
10 |
-1 |
174 |
537.456 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.18 |
5.09 |
-83.72 |
4 |
10 |
-1 |
174 |
537.456 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABI-1-B |
Enoyl-[acyl-carrier-protein] Reductase (cluster #1 Of 2), Bacterial |
Bacteria |
7050 |
0.34 |
Binding ≤ 10μM
|
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
4300 |
0.36 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1660 |
0.39 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.34 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9570 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CD38-1-E |
Lymphocyte Differentiation Antigen CD38 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8200 |
0.34 |
Binding ≤ 10μM |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.34 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.40 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.38 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.41 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1450 |
0.39 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
890 |
0.40 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3370 |
0.36 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
56 |
0.48 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
3800 |
0.36 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9600 |
0.33 |
Functional ≤ 10μM
|
|
Z50607-4-O |
Human Immunodeficiency Virus 1 (cluster #4 Of 10), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM |
|
Z50652-3-O |
Influenza A Virus (cluster #3 Of 4), Other |
Other |
6820 |
0.34 |
Functional ≤ 10μM
|
|
Z80150-2-O |
H9c2 (Cardiomyoblast Cells) (cluster #2 Of 2), Other |
Other |
5530 |
0.35 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
7500 |
0.34 |
Functional ≤ 10μM
|
|
Z80420-1-O |
RBL-2H3 (Basophilic Leukemia Cells) (cluster #1 Of 2), Other |
Other |
5800 |
0.35 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
800 |
0.41 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
79 |
0.47 |
Binding ≤ 1μM
|
|
TOP1_RAT |
Q9WUL0
|
DNA Topoisomerase 1, Rat |
660 |
0.41 |
Binding ≤ 1μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
800 |
0.41 |
Binding ≤ 1μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
800 |
0.41 |
Binding ≤ 1μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
850 |
0.40 |
Binding ≤ 1μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
550 |
0.42 |
Binding ≤ 1μM
|
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
1800 |
0.38 |
Binding ≤ 10μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
1550 |
0.39 |
Binding ≤ 10μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
9570 |
0.33 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1540 |
0.39 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
8900 |
0.34 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
6200 |
0.35 |
Binding ≤ 10μM
|
|
Z104294 |
Z104294
|
Cyclin-dependent Kinase 5/CDK5 Activator 1 |
3800 |
0.36 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
79 |
0.47 |
Binding ≤ 10μM
|
|
TOP1_RAT |
Q9WUL0
|
DNA Topoisomerase 1, Rat |
5000 |
0.35 |
Binding ≤ 10μM
|
|
Q965D5_PLAFA |
Q965D5
|
Enoyl-acyl-carrier Protein Reductase, Plafa |
2000 |
0.38 |
Binding ≤ 10μM
|
|
FABI_ECOLI |
P0AEK4
|
Enoyl-[acyl-carrier-protein] Reductase, Ecoli |
7050 |
0.34 |
Binding ≤ 10μM
|
|
Q965D7_PLAFA |
Q965D7
|
Fatty Acid Synthase, Plafa |
5000 |
0.35 |
Binding ≤ 10μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
6200 |
0.35 |
Binding ≤ 10μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
6200 |
0.35 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
800 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
800 |
0.41 |
Binding ≤ 10μM
|
|
LGUL_HUMAN |
Q04760
|
Glyoxalase I, Human |
7700 |
0.34 |
Binding ≤ 10μM
|
|
CD38_HUMAN |
P28907
|
Lymphocyte Differentiation Antigen CD38, Human |
8200 |
0.34 |
Binding ≤ 10μM |
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
4900 |
0.35 |
Binding ≤ 10μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
850 |
0.40 |
Binding ≤ 10μM
|
|
LOX1_SOYBN |
P08170
|
Seed Lipoxygenase-1, Soybn |
3200 |
0.37 |
Binding ≤ 10μM
|
|
NANH_CLOPE |
P10481
|
Sialidase, Clope |
4300 |
0.36 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
550 |
0.42 |
Binding ≤ 10μM
|
|
XDH_BOVIN |
P80457
|
Xanthine Dehydrogenase, Bovin |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
1450 |
0.39 |
Functional ≤ 10μM
|
|
Z80150 |
Z80150
|
H9c2 (Cardiomyoblast Cells) |
5530 |
0.35 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
10000 |
0.33 |
Functional ≤ 10μM
|
|
Z50652 |
Z50652
|
Influenza A Virus |
6820 |
0.34 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
9600 |
0.33 |
Functional ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
7390 |
0.34 |
Functional ≤ 10μM
|
|
Z80420 |
Z80420
|
RBL-2H3 (Basophilic Leukemia Cells) |
3700 |
0.36 |
Functional ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
1249 |
0.39 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
3370 |
0.36 |
ADME/T ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
56 |
0.48 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
-0.94 |
-14.61 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
12.2 |
-15.48 |
2 |
6 |
0 |
85 |
444.531 |
11 |
↓
|
|
|
|
Analogs
-
34583707
-
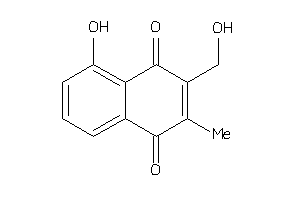
-
39283830
-
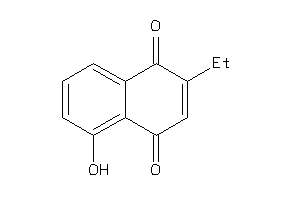
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
270 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
4.24 |
-7.85 |
1 |
3 |
0 |
54 |
188.182 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
2.83 |
-10.23 |
2 |
4 |
0 |
71 |
228.203 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.80 |
3.56 |
-58.09 |
1 |
4 |
-1 |
73 |
227.195 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NFKB1-1-E |
Nuclear Factor NF-kappa-B P105 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.55 |
Binding ≤ 10μM
|
|
CP2A6-3-E |
Cytochrome P450 2A6 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
6.3 |
-12.66 |
0 |
3 |
0 |
43 |
186.166 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
3350 |
0.48 |
Functional ≤ 10μM
|
|
Z80164-6-O |
HT-1080 (Fibrosarcoma Cells) (cluster #6 Of 6), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
5.76 |
-10.08 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
-0.17 |
-14.48 |
0 |
4 |
0 |
48 |
275.263 |
0 |
↓
|
|
|
|
Analogs
-
39289597
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
0.07 |
-9.75 |
1 |
3 |
0 |
46 |
164.16 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
3.72 |
-9.73 |
3 |
4 |
0 |
74 |
310.349 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.18 |
4.48 |
-48.99 |
2 |
4 |
-1 |
77 |
309.341 |
3 |
↓
|
|
|
|
Analogs
-
39382015
-
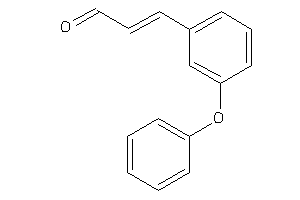
-
1529491
-
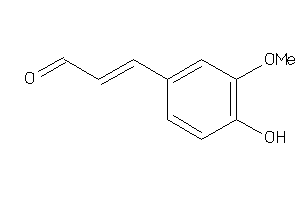
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
2.31 |
-11.21 |
0 |
3 |
0 |
35 |
192.214 |
4 |
↓
|
|
|
|
Analogs
-
5357845
-
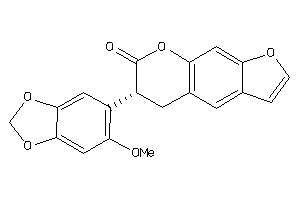
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
2.44 |
-12.93 |
0 |
6 |
0 |
67 |
338.315 |
2 |
↓
|
|
|
|
Analogs
-
5998737
-
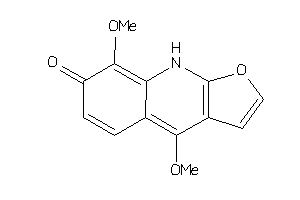
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
2.82 |
-14.24 |
0 |
5 |
0 |
54 |
259.261 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
3.88 |
-24.01 |
1 |
5 |
1 |
55 |
260.269 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
7.92 |
-12.72 |
0 |
6 |
0 |
59 |
329.352 |
4 |
↓
|
|
|
|
Analogs
-
3641069
-
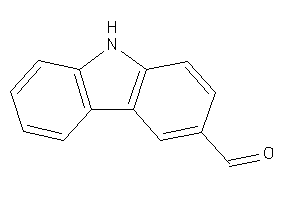
-
34540154
-
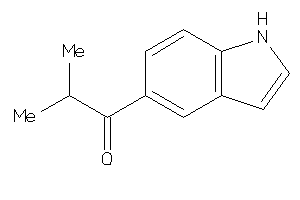
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
4.26 |
-8.56 |
1 |
2 |
0 |
33 |
145.161 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
8900 |
0.35 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
8900 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-0.85 |
-12.09 |
0 |
6 |
0 |
62 |
273.244 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.72 |
-0.14 |
-25.23 |
1 |
6 |
1 |
64 |
274.252 |
2 |
↓
|
|
|
|
Analogs
-
4227090
-
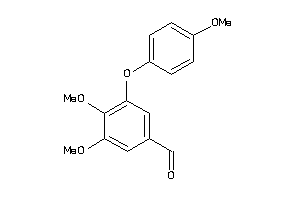
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
1.27 |
-10.7 |
1 |
4 |
0 |
56 |
182.175 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
3.85 |
-8.82 |
2 |
3 |
0 |
49 |
225.332 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
5.95 |
-9.19 |
1 |
7 |
0 |
83 |
410.422 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
7.83 |
-14.57 |
0 |
6 |
0 |
71 |
336.299 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.76 |
11.34 |
-2.7 |
1 |
1 |
0 |
20 |
296.539 |
13 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
2.43 |
-15.01 |
2 |
4 |
0 |
75 |
266.337 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7580 |
0.36 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.37 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6670 |
0.36 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
BGLR-1-E |
Beta-glucuronidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.53 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4290 |
0.38 |
Binding ≤ 10μM
|
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.43 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
660 |
0.43 |
Binding ≤ 10μM
|
|
ESR2-4-E |
Estrogen Receptor Beta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
660 |
0.43 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GBRA6-6-E |
GABA Receptor Alpha-6 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.39 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.39 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1130 |
0.42 |
Binding ≤ 10μM
|
|
P90584-1-E |
Protein Kinase Pfmrk (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.43 |
Binding ≤ 10μM
|
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.37 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
427 |
0.45 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
795 |
0.43 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
64 |
0.50 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
1600 |
0.41 |
Binding ≤ 10μM
|
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
770 |
0.43 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3400 |
0.38 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
9000 |
0.35 |
Functional ≤ 10μM |
|
Z50652-3-O |
Influenza A Virus (cluster #3 Of 4), Other |
Other |
4740 |
0.37 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
7700 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
740 |
0.43 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1000 |
0.42 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
25 |
0.53 |
Binding ≤ 1μM
|
|
DHB1_HUMAN |
P14061
|
Estradiol 17-beta-dehydrogenase 1, Human |
710 |
0.43 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
660 |
0.43 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
130 |
0.48 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
770 |
0.43 |
Binding ≤ 1μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
700 |
0.43 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
3000 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
7580 |
0.36 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
6670 |
0.36 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
3100 |
0.39 |
Binding ≤ 10μM
|
|
BGLR_RAT |
P06760
|
Beta-glucuronidase, Rat |
2800 |
0.39 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
10000 |
0.35 |
Binding ≤ 10μM
|
|
CSK2B_HUMAN |
P67870
|
Casein Kinase II Beta, Human |
10000 |
0.35 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
4000 |
0.38 |
Binding ≤ 10μM
|
|
Z104294 |
Z104294
|
Cyclin-dependent Kinase 5/CDK5 Activator 1 |
1600 |
0.41 |
Binding ≤ 10μM
|
|
CDK6_HUMAN |
Q00534
|
Cyclin-dependent Kinase 6, Human |
1700 |
0.40 |
Binding ≤ 10μM
|
|
PGH2_HUMAN |
P35354
|
Cyclooxygenase-2, Human |
8000 |
0.36 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1000 |
0.42 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
25 |
0.53 |
Binding ≤ 10μM
|
|
DHB1_HUMAN |
P14061
|
Estradiol 17-beta-dehydrogenase 1, Human |
710 |
0.43 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
1253 |
0.41 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
130 |
0.48 |
Binding ≤ 10μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
4000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
4000 |
0.38 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
3019.95172 |
0.39 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
770 |
0.43 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
1700 |
0.40 |
Binding ≤ 10μM
|
|
MRP1_HUMAN |
P33527
|
Multidrug Resistance-associated Protein 1, Human |
2400 |
0.39 |
Binding ≤ 10μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
1130 |
0.42 |
Binding ≤ 10μM
|
|
P90584_PLAFA |
P90584
|
Protein Kinase Pfmrk, Plafa |
7000 |
0.36 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
700 |
0.43 |
Binding ≤ 10μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
5200 |
0.37 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
9000 |
0.35 |
Functional ≤ 10μM
|
|
Z50652 |
Z50652
|
Influenza A Virus |
4740 |
0.37 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
3400 |
0.38 |
Functional ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
500 |
0.44 |
Functional ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
390 |
0.45 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
795 |
0.43 |
ADME/T ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
64 |
0.50 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
1.12 |
-12.82 |
3 |
5 |
0 |
91 |
270.24 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.72 |
1.4 |
-45.92 |
2 |
5 |
-1 |
94 |
269.232 |
1 |
↓
|
|