|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.40 |
Binding ≤ 10μM |
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Binding ≤ 10μM |
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM |
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM |
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.34 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4550 |
0.29 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.41 |
Binding ≤ 10μM |
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
21 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM |
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
35 |
0.40 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9XYC7_PLAFA |
Q9XYC7
|
Histone Deacetylase, Plafa |
1.8 |
0.47 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
1 |
0.48 |
Binding ≤ 1μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
31 |
0.40 |
Binding ≤ 1μM |
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
3.5 |
0.46 |
Binding ≤ 1μM |
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
0.65 |
0.49 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
1.1 |
0.48 |
Binding ≤ 1μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
0.6 |
0.50 |
Binding ≤ 1μM |
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
0.7 |
0.49 |
Binding ≤ 1μM |
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
0.7 |
0.49 |
Binding ≤ 1μM |
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
2.3 |
0.47 |
Binding ≤ 1μM |
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
105 |
0.38 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
1.2 |
0.48 |
Binding ≤ 1μM |
|
Q9XYC7_PLAFA |
Q9XYC7
|
Histone Deacetylase, Plafa |
1.8 |
0.47 |
Binding ≤ 10μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
1 |
0.48 |
Binding ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
31 |
0.40 |
Binding ≤ 10μM |
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
3.5 |
0.46 |
Binding ≤ 10μM |
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
0.65 |
0.49 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
1.1 |
0.48 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
0.6 |
0.50 |
Binding ≤ 10μM |
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
0.7 |
0.49 |
Binding ≤ 10μM |
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
0.7 |
0.49 |
Binding ≤ 10μM |
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
2.3 |
0.47 |
Binding ≤ 10μM |
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
105 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
1.2 |
0.48 |
Binding ≤ 10μM |
|
Z81034 |
Z81034
|
A2780 (Ovarian Carcinoma Cells) |
35 |
0.40 |
Functional ≤ 10μM |
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
48 |
0.39 |
Functional ≤ 10μM |
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
24 |
0.41 |
Functional ≤ 10μM |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
21 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
6.33 |
-22.79 |
4 |
5 |
0 |
77 |
349.434 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
3.37 |
5.51 |
-52.21 |
5 |
5 |
1 |
85 |
350.442 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.19 |
7.58 |
-62.39 |
3 |
5 |
-1 |
80 |
348.426 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
3.85 |
-14.84 |
3 |
11 |
0 |
148 |
532.619 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1960 |
0.27 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
SC5A4-1-E |
Low Affinity Sodium-glucose Cotransporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
-1.04 |
-11.37 |
4 |
7 |
0 |
109 |
436.888 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
4.92 |
-35.29 |
1 |
9 |
0 |
121 |
371.426 |
5 |
↓
|
|
|
|
Analogs
-
3964523
-
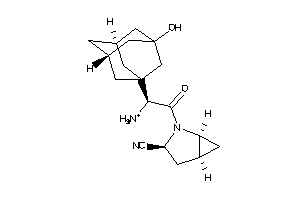
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.91 |
-12.9 |
3 |
5 |
0 |
90 |
315.417 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
11.6 |
-12.09 |
2 |
8 |
0 |
99 |
440.507 |
5 |
↓
|
|
|
|
Analogs
-
35793633
-
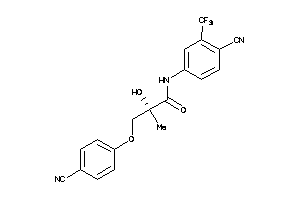
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
6.29 |
-13.9 |
2 |
6 |
0 |
106 |
389.333 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
3.12 |
4.24 |
-13.8 |
2 |
6 |
0 |
110 |
389.333 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
11.17 |
-21.58 |
1 |
6 |
0 |
76 |
464.444 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
15.64 |
-27.12 |
0 |
1 |
1 |
4 |
290.515 |
13 |
↓
|
|
|
|
Analogs
-
8214651
-
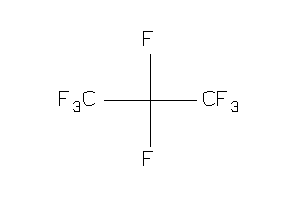
-
60255944
-
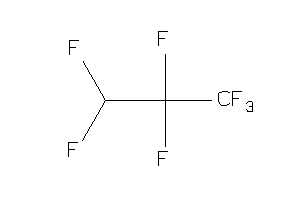
Draw
Identity
99%
90%
80%
70%
Popular Name:
1,1,1,2,3,3,3-Heptafluoropropane
1,1,1,2,3,3,3-Heptafluoropropane
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
5.65 |
-3.2 |
0 |
0 |
0 |
0 |
170.027 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
1.17 |
-11.77 |
4 |
5 |
0 |
90 |
404.459 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.15 |
11.67 |
-5.05 |
0 |
2 |
0 |
26 |
330.321 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.15 |
11.69 |
-4.55 |
0 |
2 |
0 |
26 |
330.321 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.15 |
11.7 |
-4.58 |
0 |
2 |
0 |
26 |
330.321 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.15 |
11.67 |
-5.05 |
0 |
2 |
0 |
26 |
330.321 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.32 |
Functional ≤ 10μM |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.97 |
-19.98 |
0 |
8 |
0 |
80 |
450.93 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3990 |
0.25 |
Binding ≤ 10μM
|
|
SC5A2-1-E |
Sodium/glucose Cotransporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
2.55 |
-11.17 |
4 |
6 |
0 |
99 |
434.554 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
10.12 |
-12.35 |
0 |
6 |
0 |
69 |
439.313 |
5 |
↓
|
|
|
|
Analogs
-
26506755
-
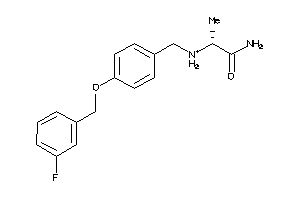
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
80 |
0.45 |
Binding ≤ 10μM
|
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
20 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB_HUMAN |
P27338
|
Monoamine Oxidase B, Human |
450 |
0.40 |
Binding ≤ 1μM
|
|
AOFB_RAT |
P19643
|
Monoamine Oxidase B, Rat |
98 |
0.45 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
19.7 |
0.49 |
Binding ≤ 1μM
|
|
AOFB_RAT |
P19643
|
Monoamine Oxidase B, Rat |
98 |
0.45 |
Binding ≤ 10μM
|
|
AOFB_HUMAN |
P27338
|
Monoamine Oxidase B, Human |
450 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
19.7 |
0.49 |
Binding ≤ 10μM
|
|
SCN1A_RAT |
P04774
|
Sodium Channel Protein Type I Alpha Subunit, Rat |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A_RAT |
P04775
|
Sodium Channel Protein Type II Alpha Subunit, Rat |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A_RAT |
P08104
|
Sodium Channel Protein Type III Alpha Subunit, Rat |
8200 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A_RAT |
O88420
|
Sodium Channel Protein Type VIII Alpha Subunit, Rat |
8200 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
4.1 |
-11.5 |
3 |
4 |
0 |
64 |
302.349 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
0.06 |
-12.38 |
2 |
9 |
0 |
107 |
454.87 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
3.82 |
5.58 |
-14.36 |
2 |
9 |
0 |
111 |
454.87 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.19 |
0.16 |
-38.89 |
3 |
9 |
1 |
108 |
455.878 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.29 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.25 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2850 |
0.22 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
670 |
0.25 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.30 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1230 |
0.24 |
Binding ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
720 |
0.25 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
390 |
0.26 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
140 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
310 |
0.26 |
Binding ≤ 1μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
670 |
0.25 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
10.2 |
0.32 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
32 |
0.30 |
Binding ≤ 1μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
3 |
0.34 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
66 |
0.29 |
Binding ≤ 1μM
|
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
28 |
0.30 |
Binding ≤ 1μM
|
|
PRKDC_HUMAN |
P78527
|
DNA-dependent Protein Kinase, Human |
1230 |
0.24 |
Binding ≤ 10μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
310 |
0.26 |
Binding ≤ 10μM
|
|
NTRK1_HUMAN |
P04629
|
Nerve Growth Factor Receptor Trk-A, Human |
2850 |
0.22 |
Binding ≤ 10μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
670 |
0.25 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
10.2 |
0.32 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
32 |
0.30 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
3 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
66 |
0.29 |
Binding ≤ 10μM
|
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
28 |
0.30 |
Binding ≤ 10μM
|
|
Z81034 |
Z81034
|
A2780 (Ovarian Carcinoma Cells) |
140 |
0.27 |
Functional ≤ 10μM
|
|
Z80240 |
Z80240
|
MDA-MB-361 |
720 |
0.25 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
280 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
5.07 |
-19.39 |
1 |
10 |
0 |
108 |
513.649 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.44 |
7.29 |
-60.13 |
2 |
10 |
1 |
109 |
514.657 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A9-1-E |
Glycine Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
7.88 |
-18.77 |
0 |
7 |
0 |
80 |
543.461 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
14.32 |
-36.04 |
1 |
3 |
1 |
29 |
428.596 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
12.09 |
-13.41 |
2 |
9 |
0 |
107 |
615.403 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
3.09 |
-18.97 |
2 |
8 |
0 |
87 |
406.508 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-3.09 |
-20.68 |
1 |
6 |
0 |
61 |
353.419 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.01 |
8.59 |
-30.68 |
0 |
7 |
1 |
78 |
363.397 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
-2.01 |
8.42 |
-30.8 |
0 |
7 |
1 |
78 |
363.397 |
5 |
↓
|
|
|
|
Analogs
-
3322
-
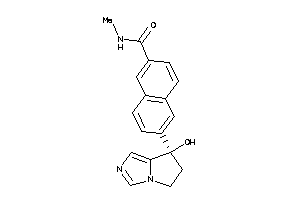
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
5.49 |
-16.29 |
2 |
5 |
0 |
67 |
307.353 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.09 |
6 |
-45.13 |
3 |
5 |
1 |
68 |
308.361 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
-0.55 |
-12.57 |
3 |
7 |
0 |
92 |
482.821 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
5.67 |
-13.37 |
3 |
7 |
0 |
96 |
364.405 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
-0.02 |
-8.72 |
1 |
3 |
0 |
38 |
388.555 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.89 |
11.4 |
-10.59 |
1 |
6 |
0 |
64 |
485.506 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.89 |
11.72 |
-38.1 |
2 |
6 |
1 |
65 |
486.514 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
38 |
0.35 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
1 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
4.68 |
-15.82 |
2 |
8 |
0 |
101 |
419.384 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.92 |
5.63 |
-58.42 |
1 |
8 |
-1 |
104 |
418.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
4.51 |
-0.35 |
1 |
1 |
0 |
12 |
167.296 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.77 |
11.95 |
-24.08 |
4 |
14 |
0 |
175 |
738.89 |
13 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.77 |
12.38 |
-49.7 |
5 |
14 |
1 |
176 |
739.898 |
13 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.77 |
12.83 |
-81.8 |
6 |
14 |
2 |
177 |
740.906 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
60 |
0.32 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
60 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
8.62 |
-12.65 |
3 |
8 |
0 |
101 |
434.544 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.19 |
9 |
-36.4 |
4 |
8 |
1 |
102 |
435.552 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.19 |
9.47 |
-88.21 |
5 |
8 |
2 |
104 |
436.56 |
9 |
↓
|
|
|
|
Analogs
-
44699816
-
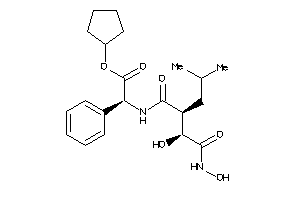
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
6.52 |
-17.31 |
4 |
8 |
0 |
125 |
406.479 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.43 |
7.75 |
-54.64 |
3 |
8 |
-1 |
128 |
405.471 |
10 |
↓
|
|
|
|
Analogs
-
38460704
-
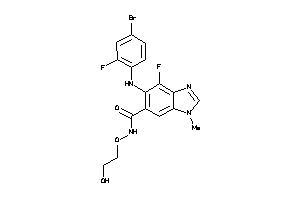
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
6.81 |
-25.34 |
3 |
7 |
0 |
88 |
457.687 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
3.49 |
5.24 |
-13.42 |
3 |
7 |
0 |
92 |
457.687 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.42 |
7.25 |
-59.59 |
4 |
7 |
1 |
90 |
458.695 |
6 |
↓
|
|
|
|
Analogs
-
40914223
-
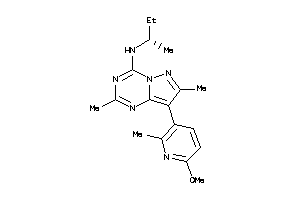
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2660 |
0.31 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM |
|
NK2R-1-E |
Neurokinin 2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
129 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
9.42 |
-9 |
1 |
7 |
0 |
77 |
340.431 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.69 |
-11.53 |
1 |
5 |
0 |
64 |
389.842 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102352-1-O |
Plasma (cluster #1 Of 3), Other |
Other |
6400 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
-4.01 |
-21.95 |
1 |
8 |
0 |
101 |
496.633 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SHH-1-E |
Sonic Hedgehog Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.27 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.07 |
-22.4 |
1 |
5 |
0 |
76 |
421.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.42 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
63 |
0.42 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1005 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
5.59 |
-11.75 |
4 |
8 |
0 |
122 |
321.344 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.15 |
-12.39 |
4 |
6 |
0 |
99 |
386.444 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
5.88 |
-19.7 |
1 |
8 |
0 |
78 |
436.512 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
8.15 |
-53.63 |
2 |
8 |
1 |
79 |
437.52 |
8 |
↓
|
|
|
|
Analogs
-
3965914
-
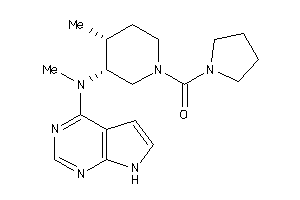
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DCLK3-1-E |
Serine/threonine-protein Kinase DCLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
JAK1-1-E |
Tyrosine-protein Kinase JAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM |
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM |
|
KCC1A-1-E |
CaM Kinase I Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3870 |
0.33 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.39 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
ROCK2-2-E |
Rho-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.33 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.33 |
Binding ≤ 10μM
|
|
TNK1-1-E |
Non-receptor Tyrosine-protein Kinase TNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.38 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.38 |
Binding ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
570 |
0.38 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
7840 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TNK1_HUMAN |
Q13470
|
Non-receptor Tyrosine-protein Kinase TNK1, Human |
640 |
0.38 |
Binding ≤ 1μM
|
|
NUAK2_HUMAN |
Q9H093
|
NUAK Family SNF1-like Kinase 2, Human |
420 |
0.39 |
Binding ≤ 1μM
|
|
PKN1_HUMAN |
Q16512
|
Protein Kinase N1, Human |
200 |
0.41 |
Binding ≤ 1μM
|
|
DCLK3_HUMAN |
Q9C098
|
Serine/threonine-protein Kinase DCLK3, Human |
4.5 |
0.51 |
Binding ≤ 1μM
|
|
JAK1_HUMAN |
P23458
|
Tyrosine-protein Kinase JAK1, Human |
1.7 |
0.53 |
Binding ≤ 1μM
|
|
JAK2_HUMAN |
O60674
|
Tyrosine-protein Kinase JAK2, Human |
1.6 |
0.54 |
Binding ≤ 1μM
|
|
JAK3_MOUSE |
Q62137
|
Tyrosine-protein Kinase JAK3, Mouse |
48 |
0.45 |
Binding ≤ 1μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
0.7 |
0.56 |
Binding ≤ 1μM
|
|
TYK2_HUMAN |
P29597
|
Tyrosine-protein Kinase TYK2, Human |
176 |
0.41 |
Binding ≤ 1μM
|
|
KCC1A_HUMAN |
Q14012
|
CaM Kinase I Alpha, Human |
5000 |
0.32 |
Binding ≤ 10μM
|
|
TNK1_HUMAN |
Q13470
|
Non-receptor Tyrosine-protein Kinase TNK1, Human |
640 |
0.38 |
Binding ≤ 10μM
|
|
NUAK2_HUMAN |
Q9H093
|
NUAK Family SNF1-like Kinase 2, Human |
420 |
0.39 |
Binding ≤ 10μM
|
|
PKN1_HUMAN |
Q16512
|
Protein Kinase N1, Human |
200 |
0.41 |
Binding ≤ 10μM
|
|
ROCK2_HUMAN |
O75116
|
Rho-associated Protein Kinase 2, Human |
3400 |
0.33 |
Binding ≤ 10μM
|
|
KS6A2_HUMAN |
Q15349
|
Ribosomal Protein S6 Kinase Alpha 2, Human |
1400 |
0.36 |
Binding ≤ 10μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
1200 |
0.36 |
Binding ≤ 10μM
|
|
DCLK3_HUMAN |
Q9C098
|
Serine/threonine-protein Kinase DCLK3, Human |
4.5 |
0.51 |
Binding ≤ 10μM
|
|
STK3_HUMAN |
Q13188
|
Serine/threonine-protein Kinase MST2, Human |
4300 |
0.33 |
Binding ≤ 10μM
|
|
JAK1_HUMAN |
P23458
|
Tyrosine-protein Kinase JAK1, Human |
1.7 |
0.53 |
Binding ≤ 10μM
|
|
JAK2_HUMAN |
O60674
|
Tyrosine-protein Kinase JAK2, Human |
1.6 |
0.54 |
Binding ≤ 10μM
|
|
JAK3_MOUSE |
Q62137
|
Tyrosine-protein Kinase JAK3, Mouse |
48 |
0.45 |
Binding ≤ 10μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
0.7 |
0.56 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
1800 |
0.35 |
Binding ≤ 10μM
|
|
TYK2_HUMAN |
P29597
|
Tyrosine-protein Kinase TYK2, Human |
176 |
0.41 |
Binding ≤ 10μM
|
|
Z80036 |
Z80036
|
BaF3 (IL-3-dependent Pro-B-cells) |
1574 |
0.35 |
Functional ≤ 10μM
|
|
Z81072 |
Z81072
|
Jurkat (Acute Leukemic T-cells) |
7840 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
9.01 |
-25.29 |
1 |
7 |
0 |
89 |
312.377 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.01 |
6.74 |
-49.17 |
1 |
7 |
1 |
86 |
313.385 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.45 |
9.48 |
-43.37 |
2 |
7 |
1 |
90 |
313.385 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JAK1-1-E |
Tyrosine-protein Kinase JAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.39 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
9.25 |
-17.45 |
1 |
6 |
0 |
83 |
306.373 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1998 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
3.57 |
-12.57 |
2 |
6 |
0 |
97 |
366.428 |
5 |
↓
|
|