|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.18 |
-0.81 |
-48.19 |
3 |
6 |
-1 |
110 |
218.229 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.18 |
-0.92 |
-48.11 |
3 |
6 |
-1 |
110 |
218.229 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
270 |
1.02 |
Binding ≤ 10μM
|
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
29 |
1.17 |
Functional ≤ 10μM |
|
Z50588-6-O |
Canis Familiaris (cluster #6 Of 7), Other |
Other |
2700 |
0.87 |
Functional ≤ 10μM
|
|
Z50597-8-O |
Rattus Norvegicus (cluster #8 Of 12), Other |
Other |
960 |
0.94 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
3.29 |
-45.83 |
0 |
3 |
-1 |
53 |
122.103 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.27 |
3.74 |
-46.83 |
1 |
3 |
0 |
54 |
123.111 |
1 |
↓
|
|
|
|
Analogs
-
1529313
-
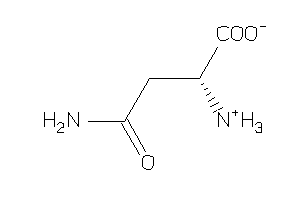
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.81 |
-5.19 |
-36.13 |
5 |
5 |
0 |
110 |
132.119 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.24 |
2.02 |
-9.53 |
3 |
5 |
0 |
80 |
135.13 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.72 |
2.43 |
-35.08 |
2 |
3 |
0 |
57 |
115.132 |
1 |
↓
|
|
|
|
Analogs
-
4806450
-
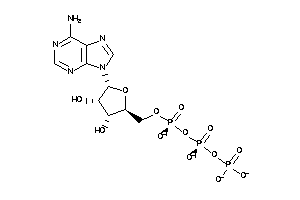
-
4806446
-
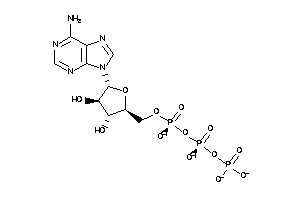
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.54 |
-2.46 |
-364.46 |
4 |
18 |
-4 |
290 |
503.15 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
-3.54 |
-3.61 |
-227.01 |
5 |
18 |
-3 |
288 |
504.158 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.54 |
-3.61 |
-227.24 |
5 |
18 |
-3 |
288 |
504.158 |
8 |
↓
|
|
|
|
Analogs
-
1532129
-
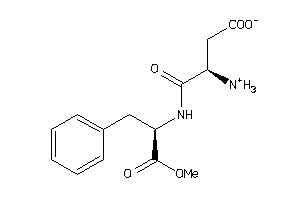
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.55 |
4.37 |
-37.85 |
4 |
7 |
0 |
123 |
294.307 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.55 |
4.05 |
-50.28 |
3 |
7 |
-1 |
122 |
293.299 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.41 |
2.19 |
-37.86 |
3 |
3 |
0 |
68 |
131.175 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
164935
-
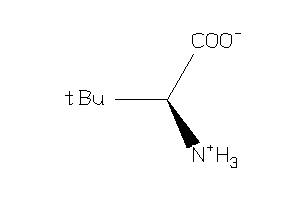
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.91 |
-1.67 |
-41.78 |
3 |
3 |
0 |
67 |
117.148 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.69 |
0.21 |
-39.21 |
3 |
3 |
0 |
68 |
89.094 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.69 |
-0.11 |
-43.06 |
2 |
3 |
-1 |
66 |
88.086 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.37 |
4.29 |
-128.65 |
5 |
13 |
-2 |
219 |
439.388 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.91 |
2.11 |
-174.25 |
4 |
13 |
-3 |
222 |
438.38 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.37 |
2.32 |
-71.85 |
6 |
13 |
-1 |
216 |
440.396 |
9 |
↓
|
|
|
|
Analogs
-
2389623
-
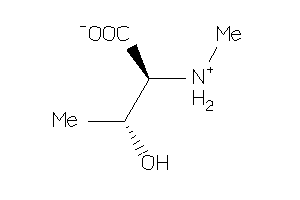
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.30 |
-2.11 |
-39.77 |
4 |
4 |
0 |
88 |
119.12 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.30 |
-2.39 |
-45.14 |
3 |
4 |
-1 |
86 |
118.112 |
2 |
↓
|
|
|
|
Analogs
-
2166829
-
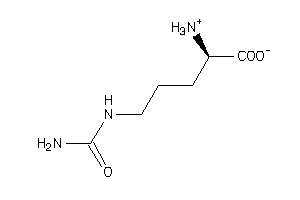
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.43 |
-2.65 |
-49.6 |
6 |
6 |
0 |
123 |
175.188 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.07 |
5.73 |
-3.36 |
0 |
1 |
0 |
12 |
175.275 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.71 |
-3.15 |
-36.35 |
3 |
3 |
0 |
67 |
121.161 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.71 |
-4.11 |
-53.68 |
3 |
3 |
-1 |
67 |
120.153 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
4.08 |
-32.56 |
4 |
4 |
0 |
84 |
204.229 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.08 |
3.78 |
-48.1 |
3 |
4 |
-1 |
82 |
203.221 |
3 |
↓
|
|
|
|
Analogs
-
388396
-
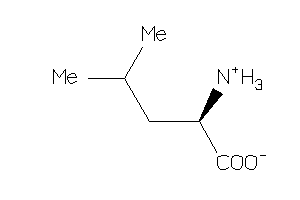
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.38 |
2.28 |
-37.79 |
3 |
3 |
0 |
68 |
131.175 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.25 |
-1.63 |
-41.1 |
4 |
5 |
0 |
94 |
131.135 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-2-E |
Glycine Receptor Subunit Alpha-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Binding ≤ 10μM
|
|
GLRA2-2-E |
Glycine Receptor Subunit Alpha-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Binding ≤ 10μM
|
|
GLRA3-2-E |
Glycine Receptor Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Binding ≤ 10μM
|
|
GLRA4-2-E |
Glycine Receptor Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Binding ≤ 10μM
|
|
GLRB-2-E |
Glycine Receptor Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
6200 |
1.46 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
86 |
1.98 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.55 |
-0.44 |
-40.12 |
3 |
3 |
0 |
68 |
75.067 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.55 |
-0.82 |
-42.26 |
2 |
3 |
-1 |
66 |
74.059 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSTK1-1-E |
Glutathione S-transferase Kappa 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.97 |
-0.38 |
-79.69 |
5 |
9 |
-1 |
166 |
306.32 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.97 |
-0.7 |
-93.89 |
4 |
9 |
-2 |
164 |
305.312 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
6.24 |
-93.26 |
7 |
7 |
2 |
113 |
440.498 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
613 |
0.87 |
Binding ≤ 10μM
|
|
EAA3-2-E |
Excitatory Amino Acid Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
51 |
1.02 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1360 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.87 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.77 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.79 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4100 |
0.75 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Binding ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.77 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
450 |
0.89 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6160 |
0.73 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.73 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3660 |
0.76 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.70 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.71 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.71 |
Functional ≤ 10μM
|
|
GRM4-1-E |
Metabotropic Glutamate Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.73 |
Functional ≤ 10μM
|
|
GRM5-5-E |
Metabotropic Glutamate Receptor 5 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.70 |
Functional ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
870 |
0.85 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 5), Other |
Other |
170 |
0.95 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
1300 |
0.82 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
613 |
0.87 |
Binding ≤ 1μM
|
|
EAA3_HUMAN |
P43005
|
Excitatory Amino Acid Transporter 3, Human |
51 |
1.02 |
Binding ≤ 1μM
|
|
NMDZ1_RAT |
P35439
|
Glutamate (NMDA) Receptor Subunit Zeta 1, Rat |
1000 |
0.84 |
Binding ≤ 1μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
1000 |
0.84 |
Binding ≤ 1μM
|
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
138.038426 |
0.96 |
Binding ≤ 1μM
|
|
GRIK1_HUMAN |
P39086
|
Glutamate Receptor Ionotropic Kainate 1, Human |
701 |
0.86 |
Binding ≤ 1μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
270 |
0.92 |
Binding ≤ 1μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
270 |
0.92 |
Binding ≤ 1μM
|
|
GRIK3_HUMAN |
Q13003
|
Glutamate Receptor Ionotropic Kainate 3, Human |
789 |
0.85 |
Binding ≤ 1μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
270 |
0.92 |
Binding ≤ 1μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
270 |
0.92 |
Binding ≤ 1μM
|
|
GRIK5_HUMAN |
Q16478
|
Glutamate Receptor Ionotropic Kainate 5, Human |
701 |
0.86 |
Binding ≤ 1μM
|
|
GRIA1_RAT |
P19490
|
Glutamate Receptor Ionotropic, AMPA 1, Rat |
169 |
0.95 |
Binding ≤ 1μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
280 |
0.92 |
Binding ≤ 1μM
|
|
GRIA2_HUMAN |
P42262
|
Glutamate Receptor Ionotropic, AMPA 2, Human |
940 |
0.84 |
Binding ≤ 1μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
249 |
0.92 |
Binding ≤ 1μM
|
|
GRIA4_HUMAN |
P48058
|
Glutamate Receptor Ionotropic, AMPA 4, Human |
868 |
0.85 |
Binding ≤ 1μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
354 |
0.90 |
Binding ≤ 1μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
250 |
0.92 |
Binding ≤ 1μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
390 |
0.90 |
Binding ≤ 1μM
|
|
GRM8_MOUSE |
P47743
|
Metabotropic Glutamate Receptor 8, Mouse |
22 |
1.07 |
Binding ≤ 1μM
|
|
GRM8_HUMAN |
O00222
|
Metabotropic Glutamate Receptor 8, Human |
5.7 |
1.15 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
170 |
0.95 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
613 |
0.87 |
Binding ≤ 10μM
|
|
EAA3_HUMAN |
P43005
|
Excitatory Amino Acid Transporter 3, Human |
51 |
1.02 |
Binding ≤ 10μM
|
|
NMDZ1_RAT |
P35439
|
Glutamate (NMDA) Receptor Subunit Zeta 1, Rat |
1000 |
0.84 |
Binding ≤ 10μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
1000 |
0.84 |
Binding ≤ 10μM
|
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
138.038426 |
0.96 |
Binding ≤ 10μM
|
|
GRIK1_HUMAN |
P39086
|
Glutamate Receptor Ionotropic Kainate 1, Human |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
270 |
0.92 |
Binding ≤ 10μM
|
|
GRIK2_HUMAN |
Q13002
|
Glutamate Receptor Ionotropic Kainate 2, Human |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3_HUMAN |
Q13003
|
Glutamate Receptor Ionotropic Kainate 3, Human |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
270 |
0.92 |
Binding ≤ 10μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
270 |
0.92 |
Binding ≤ 10μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
270 |
0.92 |
Binding ≤ 10μM
|
|
GRIK5_HUMAN |
Q16478
|
Glutamate Receptor Ionotropic Kainate 5, Human |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIA1_HUMAN |
P42261
|
Glutamate Receptor Ionotropic, AMPA 1, Human |
1360 |
0.82 |
Binding ≤ 10μM
|
|
GRIA1_MOUSE |
P23818
|
Glutamate Receptor Ionotropic, AMPA 1, Mouse |
1100 |
0.83 |
Binding ≤ 10μM
|
|
GRIA1_RAT |
P19490
|
Glutamate Receptor Ionotropic, AMPA 1, Rat |
169 |
0.95 |
Binding ≤ 10μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
1100 |
0.83 |
Binding ≤ 10μM
|
|
GRIA2_HUMAN |
P42262
|
Glutamate Receptor Ionotropic, AMPA 2, Human |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
1100 |
0.83 |
Binding ≤ 10μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
1100 |
0.83 |
Binding ≤ 10μM
|
|
GRIA4_HUMAN |
P48058
|
Glutamate Receptor Ionotropic, AMPA 4, Human |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
250 |
0.92 |
Binding ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
7300 |
0.72 |
Binding ≤ 10μM
|
|
GRM2_RAT |
P31421
|
Metabotropic Glutamate Receptor 2, Rat |
4000 |
0.76 |
Binding ≤ 10μM
|
|
GRM2_HUMAN |
Q14416
|
Metabotropic Glutamate Receptor 2, Human |
4700 |
0.75 |
Binding ≤ 10μM
|
|
GRM3_RAT |
P31422
|
Metabotropic Glutamate Receptor 3, Rat |
3000 |
0.77 |
Binding ≤ 10μM
|
|
GRM4_RAT |
P31423
|
Metabotropic Glutamate Receptor 4, Rat |
3200 |
0.77 |
Binding ≤ 10μM
|
|
GRM4_HUMAN |
Q14833
|
Metabotropic Glutamate Receptor 4, Human |
1400 |
0.82 |
Binding ≤ 10μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
1160 |
0.83 |
Binding ≤ 10μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
10000 |
0.70 |
Binding ≤ 10μM
|
|
GRM6_RAT |
P35349
|
Metabotropic Glutamate Receptor 6, Rat |
7600 |
0.72 |
Binding ≤ 10μM
|
|
GRM8_RAT |
P70579
|
Metabotropic Glutamate Receptor 8, Rat |
3400 |
0.77 |
Binding ≤ 10μM
|
|
GRM8_MOUSE |
P47743
|
Metabotropic Glutamate Receptor 8, Mouse |
22 |
1.07 |
Binding ≤ 10μM
|
|
GRM8_HUMAN |
O00222
|
Metabotropic Glutamate Receptor 8, Human |
5.7 |
1.15 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
170 |
0.95 |
Binding ≤ 10μM
|
|
NMDZ1_RAT |
P35439
|
Glutamate (NMDA) Receptor Subunit Zeta 1, Rat |
1800 |
0.80 |
Functional ≤ 10μM
|
|
GRIA1_RAT |
P19490
|
Glutamate Receptor Ionotropic, AMPA 1, Rat |
6400 |
0.73 |
Functional ≤ 10μM
|
|
GRIA1_HUMAN |
P42261
|
Glutamate Receptor Ionotropic, AMPA 1, Human |
2260 |
0.79 |
Functional ≤ 10μM
|
|
GRIA2_HUMAN |
P42262
|
Glutamate Receptor Ionotropic, AMPA 2, Human |
2190 |
0.79 |
Functional ≤ 10μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
1300 |
0.82 |
Functional ≤ 10μM
|
|
GRIA3_HUMAN |
P42263
|
Glutamate Receptor Ionotropic, AMPA 3, Human |
3660 |
0.76 |
Functional ≤ 10μM
|
|
NMDE1_RAT |
Q00959
|
Glutamate [NMDA] Receptor Subunit Epsilon 1, Rat |
1800 |
0.80 |
Functional ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
10000 |
0.70 |
Functional ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
4900 |
0.74 |
Functional ≤ 10μM
|
|
GRM2_HUMAN |
Q14416
|
Metabotropic Glutamate Receptor 2, Human |
8500 |
0.71 |
Functional ≤ 10μM
|
|
GRM2_RAT |
P31421
|
Metabotropic Glutamate Receptor 2, Rat |
290 |
0.92 |
Functional ≤ 10μM
|
|
GRM3_HUMAN |
Q14832
|
Metabotropic Glutamate Receptor 3, Human |
2500 |
0.78 |
Functional ≤ 10μM
|
|
GRM3_RAT |
P31422
|
Metabotropic Glutamate Receptor 3, Rat |
290 |
0.92 |
Functional ≤ 10μM
|
|
GRM4_RAT |
P31423
|
Metabotropic Glutamate Receptor 4, Rat |
6100 |
0.73 |
Functional ≤ 10μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
1200 |
0.83 |
Functional ≤ 10μM
|
|
GRM6_RAT |
P35349
|
Metabotropic Glutamate Receptor 6, Rat |
7600 |
0.72 |
Functional ≤ 10μM
|
|
GRM8_HUMAN |
O00222
|
Metabotropic Glutamate Receptor 8, Human |
9500 |
0.70 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1300 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.74 |
-73.91 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGLN1-2-E |
Egl Nine Homolog 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.97 |
Binding ≤ 10μM
|
|
Z80583-4-O |
Vero (Kidney Cells) (cluster #4 Of 7), Other |
Other |
1 |
1.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.65 |
3.54 |
-101.06 |
0 |
4 |
-2 |
80 |
116.072 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.65 |
1.56 |
-42.51 |
1 |
4 |
-1 |
77 |
117.08 |
3 |
↓
|
|
|
|
Analogs
-
1529198
-
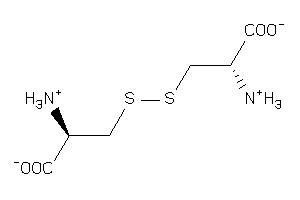
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.86 |
2.26 |
-62.06 |
6 |
6 |
0 |
136 |
240.306 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.86 |
1.97 |
-65.23 |
5 |
6 |
-1 |
134 |
239.298 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.86 |
1.67 |
-91.3 |
4 |
6 |
-2 |
132 |
238.29 |
7 |
↓
|
|
|
|
Analogs
-
2234
-
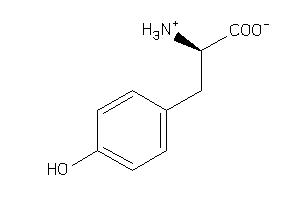
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
1.17 |
-36.43 |
4 |
4 |
0 |
88 |
181.191 |
3 |
↓
|
|
|
|
Analogs
-
1532766
-
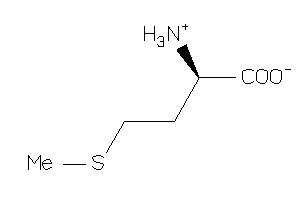
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.24 |
2.24 |
-40.54 |
3 |
3 |
0 |
68 |
149.215 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.24 |
1.91 |
-44.84 |
2 |
3 |
-1 |
66 |
148.207 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.67 |
-3.07 |
-34.14 |
4 |
4 |
0 |
88 |
105.093 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.67 |
-3.4 |
-42.43 |
3 |
4 |
-1 |
86 |
104.085 |
2 |
↓
|
|
|
|
|
|
|
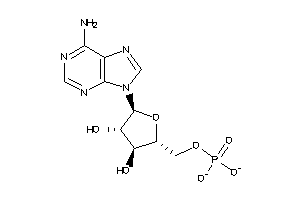
Analogs
-
3977897
-
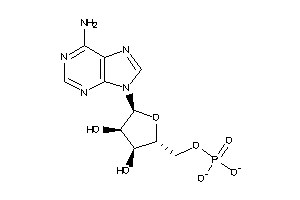
-
4806442
-
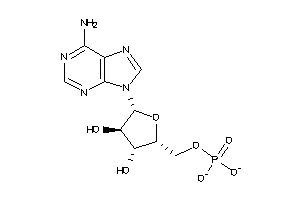
-
1532515
-
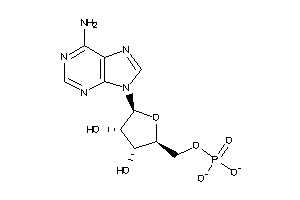
-
3201889
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
F16P1-1-E |
Fructose-1,6-bisphosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
F16P1_HUMAN |
P09467
|
Fructose-1,6-bisphosphatase, Human |
1000 |
0.37 |
Binding ≤ 1μM
|
|
Q72874_9HIV1 |
Q72874
|
Human Immunodeficiency Virus Type 1 Protease, 9hiv1 |
0.23 |
0.59 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
100 |
0.43 |
Binding ≤ 1μM
|
|
F16P1_PIG |
P00636
|
Fructose-1,6-bisphosphatase, Pig |
1300 |
0.36 |
Binding ≤ 10μM
|
|
F16P1_HUMAN |
P09467
|
Fructose-1,6-bisphosphatase, Human |
1000 |
0.37 |
Binding ≤ 10μM
|
|
Q72874_9HIV1 |
Q72874
|
Human Immunodeficiency Virus Type 1 Protease, 9hiv1 |
0.23 |
0.59 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
100 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.52 |
-2.18 |
-140.94 |
4 |
12 |
-2 |
192 |
345.208 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.52 |
-3.34 |
-50.9 |
5 |
12 |
-1 |
189 |
346.216 |
4 |
↓
|
|
|
|
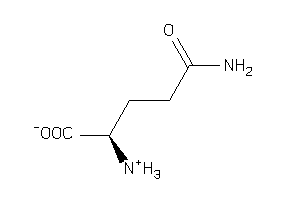
Analogs
-
1532609
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.76 |
-2.13 |
-50.63 |
5 |
5 |
0 |
111 |
146.146 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.76 |
-2.45 |
-53.15 |
4 |
5 |
-1 |
109 |
145.138 |
4 |
↓
|
|
|
|
Analogs
-
1532678
-
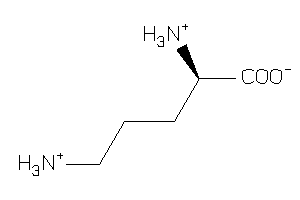
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.69 |
-0.84 |
-89.95 |
6 |
4 |
1 |
95 |
133.171 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.69 |
-1.15 |
-65.17 |
5 |
4 |
0 |
94 |
132.163 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
1638 |
0.90 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
10000 |
0.78 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.52 |
1 |
-52.73 |
3 |
5 |
-1 |
108 |
132.095 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.52 |
-1.01 |
-32.41 |
4 |
5 |
0 |
105 |
133.103 |
3 |
↓
|
|
|
|
Analogs
-
1532612
-
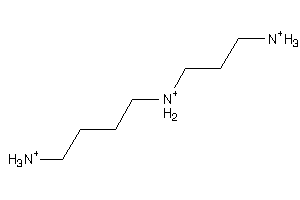
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
860 |
0.61 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
840 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
830 |
0.61 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
990 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
710 |
0.61 |
Binding ≤ 10μM
|
|
CASP2-3-E |
Caspase-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.51 |
Binding ≤ 10μM
|
|
Z100250-2-O |
Calf Thymus DNA (cluster #2 Of 2), Other |
Other |
4300 |
0.54 |
Binding ≤ 10μM
|
|
Z104302-6-O |
Glutamate NMDA Receptor (cluster #6 Of 7), Other |
Other |
5200 |
0.53 |
Binding ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
390 |
0.64 |
Functional ≤ 10μM
|
|
Z80608-2-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #2 Of 4), Other |
Other |
200 |
0.67 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
920 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
280 |
0.66 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
10 |
0.80 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
840 |
0.61 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
830 |
0.61 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
990 |
0.60 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
710 |
0.61 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
860 |
0.61 |
Binding ≤ 1μM
|
|
Z100250 |
Z100250
|
Calf Thymus DNA |
4300 |
0.54 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
840 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
830 |
0.61 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
990 |
0.60 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
710 |
0.61 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
860 |
0.61 |
Binding ≤ 10μM
|
|
CASP2_HUMAN |
P42575
|
Caspase-2, Human |
8500 |
0.51 |
Binding ≤ 10μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
5200 |
0.53 |
Binding ≤ 10μM
|
|
Z80193 |
Z80193
|
L1210 (Lymphocytic Leukemia Cells) |
390 |
0.64 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
280 |
0.66 |
Functional ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
2100 |
0.57 |
Functional ≤ 10μM
|
|
Z80608 |
Z80608
|
ZR-75-1 (Breast Carcinoma Cells) |
200 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.13 |
0.14 |
-262.68 |
10 |
4 |
4 |
88 |
206.378 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-0.62 |
-97.52 |
8 |
4 |
2 |
85 |
204.362 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-1.59 |
-88.23 |
8 |
4 |
2 |
82 |
204.362 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Functional ≤ 10μM
|
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
9700 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.12 |
-5.47 |
-13.37 |
2 |
6 |
0 |
107 |
175.116 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.12 |
-4.62 |
-15.59 |
2 |
6 |
0 |
107 |
175.116 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.93 |
-8.02 |
-141.02 |
3 |
6 |
-2 |
120 |
174.108 |
2 |
↓
|
|
|
|
Analogs
-
1532749
-
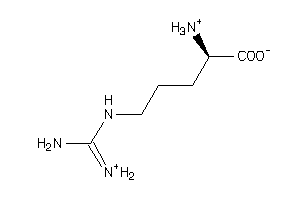
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-7-E |
Nitric Oxide Synthase, Inducible (cluster #7 Of 9), Eukaryotic |
Eukaryotes |
7000 |
0.60 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2_HUMAN |
P35228
|
Nitric Oxide Synthase, Inducible, Human |
7000 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.63 |
0.33 |
-74.97 |
8 |
6 |
1 |
131 |
175.212 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.64 |
10.11 |
-7.64 |
0 |
2 |
0 |
20 |
328.209 |
1 |
↓
|
|
|
|
Analogs
-
1532731
-
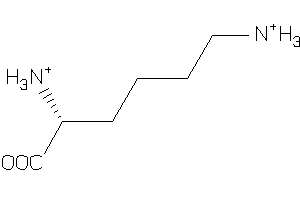
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.18 |
-0.05 |
-85.42 |
6 |
4 |
1 |
95 |
147.198 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.90 |
2.73 |
-41.42 |
0 |
3 |
-1 |
57 |
87.054 |
1 |
↓
|
|
|
|
|
|
|
|