|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.42 |
-15.43 |
1 |
5 |
0 |
68 |
206.23 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BKRB1-1-E |
Bradykinin B1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
205 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
5.11 |
-15.52 |
2 |
7 |
0 |
101 |
490.816 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
2.92 |
-11.1 |
4 |
5 |
0 |
90 |
444.524 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
11.17 |
-21.58 |
1 |
6 |
0 |
76 |
464.444 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
5.55 |
-19.27 |
2 |
10 |
0 |
121 |
531.679 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.69 |
4.05 |
-64.92 |
1 |
10 |
-1 |
124 |
530.671 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.23 |
7.86 |
-52.96 |
3 |
10 |
1 |
122 |
532.687 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
9.62 |
-15.78 |
3 |
5 |
0 |
82 |
392.499 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
9.33 |
-15.15 |
3 |
11 |
0 |
129 |
470.461 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.68 |
9.46 |
-40.06 |
4 |
11 |
1 |
130 |
471.469 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.09 |
-13.27 |
-11.12 |
6 |
6 |
0 |
121 |
182.172 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
3.77 |
-10.71 |
3 |
4 |
0 |
65 |
284.29 |
3 |
↓
|
|
|
|
Analogs
-
32500347
-
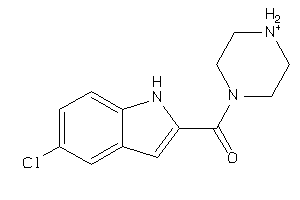
-
35527025
-
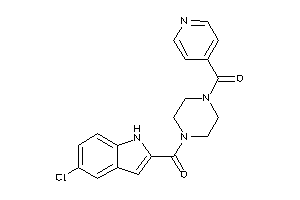
-
5348016
-
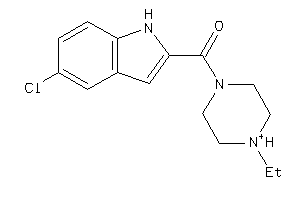
-
1491119
-
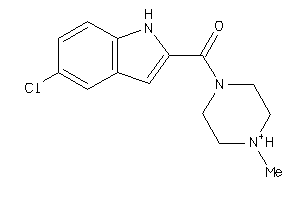
-
4704259
-
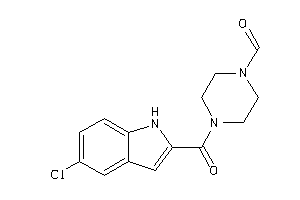
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
977.237221 |
0.44 |
Binding ≤ 1μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
12 |
0.58 |
Binding ≤ 1μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
2.4 |
0.64 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
977.237221 |
0.44 |
Binding ≤ 10μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
8511.38038 |
0.37 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
2238.72114 |
0.42 |
Binding ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
1318.25674 |
0.43 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
12 |
0.58 |
Binding ≤ 10μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
2.4 |
0.64 |
Binding ≤ 10μM
|
|
Z100489 |
Z100489
|
Eosinophils (Eosinophils) |
86 |
0.52 |
Functional ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z100488 |
Z100488
|
Mast Cells (Bone-marrow Mast Cells) |
40 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
4.89 |
-8.18 |
1 |
4 |
0 |
39 |
277.755 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
2.03 |
-12.17 |
0 |
2 |
0 |
29 |
177.294 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
2.03 |
-12.09 |
0 |
2 |
0 |
29 |
177.294 |
5 |
↓
|
|
|
|
Analogs
-
11677851
-
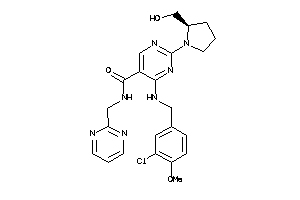
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
5.52 |
-13.88 |
3 |
10 |
0 |
129 |
483.96 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.38 |
5.96 |
-34.37 |
4 |
10 |
1 |
130 |
484.968 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.38 |
3.59 |
-35.92 |
4 |
10 |
1 |
130 |
484.968 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
MP2K1-3-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
5.85 |
-10.31 |
2 |
4 |
0 |
65 |
267.284 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
519 |
0.26 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR_HUMAN |
P30559
|
Oxytocin Receptor, Human |
519 |
0.26 |
Binding ≤ 1μM
|
|
V1AR_HUMAN |
P37288
|
Vasopressin V1a Receptor, Human |
124 |
0.28 |
Binding ≤ 1μM
|
|
V1AR_RAT |
P30560
|
Vasopressin V1a Receptor, Rat |
340 |
0.27 |
Binding ≤ 1μM
|
|
V2R_HUMAN |
P30518
|
Vasopressin V2 Receptor, Human |
1 |
0.37 |
Binding ≤ 1μM
|
|
V2R_RAT |
Q00788
|
Vasopressin V2 Receptor, Rat |
0.5 |
0.38 |
Binding ≤ 1μM
|
|
OXYR_HUMAN |
P30559
|
Oxytocin Receptor, Human |
519 |
0.26 |
Binding ≤ 10μM
|
|
V1AR_HUMAN |
P37288
|
Vasopressin V1a Receptor, Human |
124 |
0.28 |
Binding ≤ 10μM
|
|
V1AR_RAT |
P30560
|
Vasopressin V1a Receptor, Rat |
340 |
0.27 |
Binding ≤ 10μM
|
|
V2R_RAT |
Q00788
|
Vasopressin V2 Receptor, Rat |
0.5 |
0.38 |
Binding ≤ 10μM
|
|
V2R_HUMAN |
P30518
|
Vasopressin V2 Receptor, Human |
1 |
0.37 |
Binding ≤ 10μM
|
|
V2R_HUMAN |
P30518
|
Vasopressin V2 Receptor, Human |
5 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
1.8 |
-12.67 |
1 |
5 |
0 |
54 |
473.935 |
3 |
↓
|
|
|
|
Analogs
-
552049
-
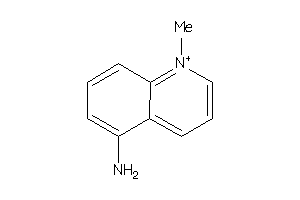
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.79 |
4.48 |
-23.02 |
4 |
3 |
1 |
56 |
224.287 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
8.91 |
-32.71 |
0 |
5 |
1 |
41 |
336.367 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.04 |
12.75 |
-23.13 |
3 |
8 |
0 |
112 |
503.591 |
6 |
↓
|
|
|
|
Analogs
-
38226010
-
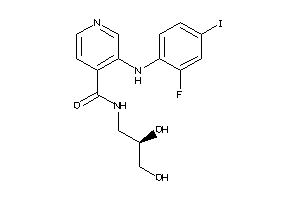
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
0.01 |
-10.57 |
4 |
6 |
0 |
94 |
431.205 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
38 |
0.35 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
1 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
4.68 |
-15.82 |
2 |
8 |
0 |
101 |
419.384 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.92 |
5.63 |
-58.42 |
1 |
8 |
-1 |
104 |
418.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
12.79 |
-10.7 |
1 |
5 |
0 |
62 |
438.474 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
10.84 |
-12.82 |
3 |
8 |
0 |
99 |
474.487 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
10.85 |
-12.66 |
3 |
8 |
0 |
99 |
474.487 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
-0.55 |
-12.57 |
3 |
7 |
0 |
92 |
482.821 |
6 |
↓
|
|
|
|
Analogs
-
15889372
-
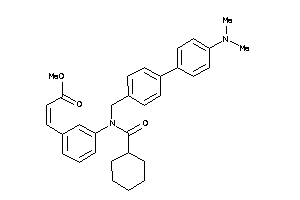
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H4-1-E |
Bile Acid Receptor FXR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.29 |
Binding ≤ 10μM
|
|
NR1H4-1-E |
Bile Acid Receptor FXR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.51 |
17.09 |
-13.12 |
0 |
5 |
0 |
50 |
496.651 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
10 |
-23.65 |
2 |
10 |
0 |
111 |
632.664 |
12 |
↓
|
|
Ref
Reference (pH 7)
|
4.57 |
7.07 |
-16.91 |
2 |
10 |
0 |
115 |
632.664 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.38 |
12.26 |
-57.4 |
3 |
10 |
1 |
112 |
633.672 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.26 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.25 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
198 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
10.31 |
-16.66 |
2 |
9 |
0 |
98 |
513.598 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.76 |
10.23 |
-3.75 |
2 |
2 |
0 |
40 |
400.647 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-3.09 |
-20.68 |
1 |
6 |
0 |
61 |
353.419 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
0.38 |
-13.78 |
1 |
7 |
0 |
90 |
443.521 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
2 |
1.11 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
36 |
0.95 |
Binding ≤ 10μM
|
|
CAH9-6-E |
Carbonic Anhydrase IX (cluster #6 Of 11), Eukaryotic |
Eukaryotes |
63 |
0.92 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
-0.12 |
-11.55 |
2 |
4 |
0 |
69 |
173.193 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.47 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.42 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.50 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
580 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
270 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1300 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCC2A_MOUSE |
P11798
|
CaM Kinase II Alpha, Mouse |
370 |
0.41 |
Binding ≤ 1μM
|
|
KCC2B_MOUSE |
P28652
|
CaM Kinase II Beta, Mouse |
370 |
0.41 |
Binding ≤ 1μM
|
|
KCC2G_MOUSE |
Q923T9
|
CaM Kinase II Gamma, Mouse |
370 |
0.41 |
Binding ≤ 1μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
49 |
0.47 |
Binding ≤ 1μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
220 |
0.42 |
Binding ≤ 1μM
|
|
PK3CA_BOVIN |
P32871
|
PI3-kinase P110-alpha Subunit, Bovin |
2.5 |
0.55 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
41 |
0.47 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
16 |
0.50 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
660 |
0.39 |
Binding ≤ 1μM
|
|
KCC2A_MOUSE |
P11798
|
CaM Kinase II Alpha, Mouse |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2B_MOUSE |
P28652
|
CaM Kinase II Beta, Mouse |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2G_MOUSE |
Q923T9
|
CaM Kinase II Gamma, Mouse |
370 |
0.41 |
Binding ≤ 10μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
3400 |
0.35 |
Binding ≤ 10μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
49 |
0.47 |
Binding ≤ 10μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
220 |
0.42 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
41 |
0.47 |
Binding ≤ 10μM
|
|
PK3CA_BOVIN |
P32871
|
PI3-kinase P110-alpha Subunit, Bovin |
2.5 |
0.55 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
16 |
0.50 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
660 |
0.39 |
Binding ≤ 10μM
|
|
Z81034 |
Z81034
|
A2780 (Ovarian Carcinoma Cells) |
270 |
0.42 |
Functional ≤ 10μM
|
|
Z103203 |
Z103203
|
A375 |
580 |
0.40 |
Functional ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
1300 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
4.03 |
-14.97 |
1 |
5 |
0 |
58 |
313.382 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.98 |
4.35 |
-28.48 |
2 |
5 |
1 |
60 |
314.39 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
7.67 |
-12.34 |
1 |
5 |
0 |
55 |
336.391 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.35 |
7.26 |
-44.91 |
0 |
5 |
-1 |
58 |
335.383 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
6.83 |
-20.73 |
1 |
8 |
0 |
91 |
351.366 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
7.83 |
-51.3 |
2 |
8 |
1 |
92 |
352.374 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
12.77 |
-16.01 |
2 |
8 |
0 |
105 |
397.442 |
3 |
↓
|
|
|
|
Analogs
-
14975141
-
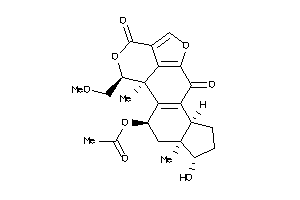
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
193 |
0.30 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10 |
0.36 |
Functional ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1464 |
0.26 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
193 |
0.30 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
0.5 |
0.42 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 1μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
0.5 |
0.42 |
Binding ≤ 1μM
|
|
P85B_HUMAN |
O00459
|
PI3-kinase P85-beta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 1μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
193 |
0.30 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
0.5 |
0.42 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
0.5 |
0.42 |
Binding ≤ 10μM
|
|
P85B_HUMAN |
O00459
|
PI3-kinase P85-beta Subunit, Human |
0.5 |
0.42 |
Binding ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
1464 |
0.26 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
10 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
1.2 |
-13.15 |
1 |
8 |
0 |
112 |
430.453 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.77 |
14.76 |
-17.99 |
0 |
6 |
0 |
77 |
469.548 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.34 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.39 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.34 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
210 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
16 |
0.36 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
44 |
0.34 |
Binding ≤ 1μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
4.6 |
0.39 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
49 |
0.34 |
Binding ≤ 1μM
|
|
MTOR_RAT |
P42346
|
Serine/threonine-protein Kinase MTOR, Rat |
45 |
0.34 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
16 |
0.36 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
44 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
4.6 |
0.39 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
49 |
0.34 |
Binding ≤ 10μM
|
|
MTOR_RAT |
P42346
|
Serine/threonine-protein Kinase MTOR, Rat |
45 |
0.34 |
Binding ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
210 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
-4.94 |
-11.62 |
0 |
9 |
0 |
81 |
417.42 |
4 |
↓
|
|
|
|
Analogs
-
19862654
-
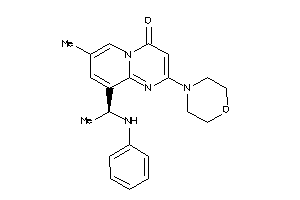
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
9.08 |
-11.61 |
1 |
6 |
0 |
59 |
364.449 |
4 |
↓
|
|
|
|
Analogs
-
19862652
-
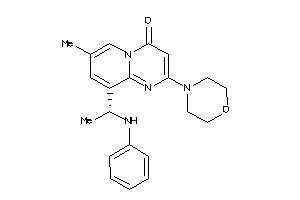
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
9.05 |
-11.56 |
1 |
6 |
0 |
59 |
364.449 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.29 |
Binding ≤ 10μM
|
|
DAPK1-1-E |
Death-associated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.30 |
Binding ≤ 10μM
|
|
DAPK2-1-E |
Death-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.33 |
Binding ≤ 10μM
|
|
LIMK2-1-E |
LIM Domain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.29 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.31 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.31 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.43 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
PK3CB-1-E |
PI3-kinase P110-beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.29 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
330 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
180 |
0.36 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
540 |
0.34 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
540 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
840 |
0.33 |
Binding ≤ 1μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
10 |
0.43 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
3.6 |
0.45 |
Binding ≤ 1μM
|
|
PK3CA_BOVIN |
P32871
|
PI3-kinase P110-alpha Subunit, Bovin |
3.6 |
0.45 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
3 |
0.46 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
250 |
0.36 |
Binding ≤ 1μM
|
|
DAPK1_HUMAN |
P53355
|
Death-associated Protein Kinase 1, Human |
2400 |
0.30 |
Binding ≤ 10μM
|
|
DAPK2_HUMAN |
Q9UIK4
|
Death-associated Protein Kinase 2, Human |
2700 |
0.30 |
Binding ≤ 10μM
|
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
840 |
0.33 |
Binding ≤ 10μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
3900 |
0.29 |
Binding ≤ 10μM
|
|
LIMK2_HUMAN |
P53671
|
LIM Domain Kinase 2, Human |
3500 |
0.29 |
Binding ≤ 10μM
|
|
MYLK_HUMAN |
Q15746
|
Myosin Light Chain Kinase, Smooth Muscle, Human |
1500 |
0.31 |
Binding ≤ 10μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
10 |
0.43 |
Binding ≤ 10μM
|
|
PK3CA_BOVIN |
P32871
|
PI3-kinase P110-alpha Subunit, Bovin |
3.6 |
0.45 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
3.6 |
0.45 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
3 |
0.46 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
250 |
0.36 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
2300 |
0.30 |
Binding ≤ 10μM
|
|
MRCKA_HUMAN |
Q5VT25
|
Serine/threonine-protein Kinase MRCK-A, Human |
1800 |
0.31 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
3700 |
0.29 |
Binding ≤ 10μM
|
|
Z103203 |
Z103203
|
A375 |
330 |
0.35 |
Functional ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
540 |
0.34 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
540 |
0.34 |
Functional ≤ 10μM
|
|
Z81057 |
Z81057
|
HUVEC (Umbilical Vein Endothelial Cells) |
0.6 |
0.50 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
130 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
-6.5 |
-13.05 |
1 |
7 |
0 |
84 |
348.362 |
2 |
↓
|
|
|
|
Analogs
-
12504504
-
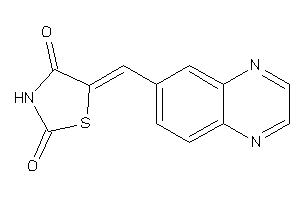
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
1.79 |
-13.38 |
1 |
5 |
0 |
76 |
257.274 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
0.08 |
-20.07 |
3 |
7 |
0 |
108 |
389.886 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
3.87 |
-11.03 |
2 |
5 |
0 |
83 |
305.286 |
2 |
↓
|
|
|
|
Analogs
-
3995884
-
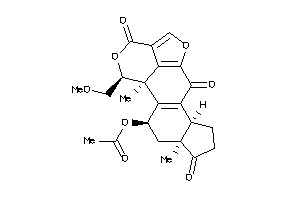
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.33 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.24 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.32 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.33 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.31 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
98 |
0.32 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRKDC_HUMAN |
P78527
|
DNA-dependent Protein Kinase, Human |
120 |
0.31 |
Binding ≤ 1μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
40 |
0.33 |
Binding ≤ 1μM
|
|
MYLK_CHICK |
P11799
|
Myosin Light Chain Kinase, Smooth Muscle, Chick |
260 |
0.30 |
Binding ≤ 1μM
|
|
MYLK_HUMAN |
Q15746
|
Myosin Light Chain Kinase, Smooth Muscle, Human |
170 |
0.31 |
Binding ≤ 1μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
1 |
0.41 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
10 |
0.36 |
Binding ≤ 1μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
4 |
0.38 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
19 |
0.35 |
Binding ≤ 1μM
|
|
P85A_HUMAN |
P27986
|
PI3-kinase P85-alpha Subunit, Human |
1.2 |
0.40 |
Binding ≤ 1μM
|
|
P85B_HUMAN |
O00459
|
PI3-kinase P85-beta Subunit, Human |
4.2 |
0.38 |
Binding ≤ 1μM
|
|
PLK1_HUMAN |
P53350
|
Serine/threonine-protein Kinase PLK1, Human |
5.8 |
0.37 |
Binding ≤ 1μM
|
|
PLK3_HUMAN |
Q9H4B4
|
Serine/threonine-protein Kinase PLK3, Human |
220 |
0.30 |
Binding ≤ 1μM
|
|
PRKDC_HUMAN |
P78527
|
DNA-dependent Protein Kinase, Human |
120 |
0.31 |
Binding ≤ 10μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
40 |
0.33 |
Binding ≤ 10μM
|
|
MYLK_CHICK |
P11799
|
Myosin Light Chain Kinase, Smooth Muscle, Chick |
260 |
0.30 |
Binding ≤ 10μM
|
|
MYLK_HUMAN |
Q15746
|
Myosin Light Chain Kinase, Smooth Muscle, Human |
170 |
0.31 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
10 |
0.36 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
4 |
0.38 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
19 |
0.35 |
Binding ≤ 10μM
|
|
P85A_HUMAN |
P27986
|
PI3-kinase P85-alpha Subunit, Human |
1.2 |
0.40 |
Binding ≤ 10μM
|
|
P85B_HUMAN |
O00459
|
PI3-kinase P85-beta Subunit, Human |
4.2 |
0.38 |
Binding ≤ 10μM
|
|
PLK1_HUMAN |
P53350
|
Serine/threonine-protein Kinase PLK1, Human |
1300 |
0.27 |
Binding ≤ 10μM
|
|
PLK3_HUMAN |
Q9H4B4
|
Serine/threonine-protein Kinase PLK3, Human |
220 |
0.30 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
98 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
8.68 |
-13.81 |
0 |
8 |
0 |
109 |
428.437 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1500 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7_HUMAN |
P36544
|
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit, Human |
1500 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
3.83 |
-11.69 |
2 |
7 |
0 |
86 |
311.725 |
4 |
↓
|
|