|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.38 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
10000 |
0.37 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
4.66 |
-7.46 |
2 |
4 |
0 |
58 |
252.273 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.36 |
2.13 |
-45.39 |
1 |
4 |
-1 |
65 |
251.265 |
2 |
↓
|
|
|
|
Analogs
-
39291087
-
-
39291089
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.1 |
-16.38 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
740 |
0.54 |
Binding ≤ 10μM
|
|
Q50565-1-A |
Carbonic Anhydrase (cluster #1 Of 1), Archaea |
Archaea |
5350 |
0.46 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
27 |
0.66 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
1030 |
0.52 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
39 |
0.65 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
105 |
0.61 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
25 |
0.67 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
22 |
0.67 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.64 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
25 |
0.67 |
Binding ≤ 10μM |
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
58 |
0.63 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
8000 |
0.45 |
Binding ≤ 10μM |
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
13 |
0.69 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
25 |
0.67 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
43 |
0.64 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.79 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
50 |
0.64 |
Binding ≤ 10μM
|
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.67 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.71 |
Functional ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.64 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
98 |
0.61 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
8 |
0.71 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
39.4 |
0.65 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
8 |
0.71 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
27 |
0.66 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
98.4 |
0.61 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
105 |
0.61 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
200 |
0.59 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
155 |
0.60 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
58 |
0.63 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
20 |
0.67 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
38 |
0.65 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
93 |
0.62 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
13 |
0.69 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
34 |
0.65 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
2 |
0.76 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
47 |
0.64 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
19 |
0.68 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
47 |
0.64 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
43 |
0.64 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.78 |
0.80 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
22 |
0.67 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
50 |
0.64 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
2.5 |
0.75 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
39.4 |
0.65 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
1070 |
0.52 |
Binding ≤ 10μM
|
|
Q50565_METTH |
Q50565
|
Carbonic Anhydrase, Metth |
5350 |
0.46 |
Binding ≤ 10μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
27 |
0.66 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
98.4 |
0.61 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
200 |
0.59 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
105 |
0.61 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
155 |
0.60 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
58 |
0.63 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
1200 |
0.52 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
38 |
0.65 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
5000 |
0.46 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
13 |
0.69 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
93 |
0.62 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
34 |
0.65 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
47 |
0.64 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
2 |
0.76 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
19 |
0.68 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
47 |
0.64 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
43 |
0.64 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.78 |
0.80 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
22 |
0.67 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
50 |
0.64 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
2.5 |
0.75 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
1030 |
0.52 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
25 |
0.67 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
8 |
0.71 |
Functional ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
50 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
-0.91 |
-11.89 |
2 |
5 |
0 |
82 |
258.324 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.76 |
-0.43 |
-46.1 |
1 |
5 |
-1 |
80 |
257.316 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.76 |
0 |
-24.43 |
2 |
5 |
0 |
81 |
258.324 |
3 |
↓
|
|
|
|
Analogs
-
4217364
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
60 |
0.78 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
9 |
0.87 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
12 |
0.85 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
481 |
0.68 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.68 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
54 |
0.78 |
Binding ≤ 10μM |
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
|
Q2PCB5-1-E |
Carbonic Anhydrase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5720 |
0.56 |
Binding ≤ 10μM |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
83 |
0.76 |
Binding ≤ 10μM
|
|
Q3I4V7-2-F |
Carbonic Anhydrase 2 (cluster #2 Of 4), Fungal |
Fungi |
10 |
0.86 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
40 |
0.80 |
Binding ≤ 10μM
|
|
Q6FTL6-1-F |
Carbonic Anhydrase (cluster #1 Of 1), Fungal |
Fungi |
11 |
0.86 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
60 |
0.78 |
Binding ≤ 1μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 1μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
17 |
0.84 |
Binding ≤ 1μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 1μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 10μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
1430 |
0.63 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 10μM
|
|
Q2PCB5_DICLA |
Q2PCB5
|
Carbonic Anhydrase, Dicla |
5720 |
0.56 |
Binding ≤ 10μM |
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 10μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
1050 |
0.64 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
12 |
0.85 |
Functional ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
25 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.36 |
-19.16 |
3 |
7 |
0 |
115 |
222.251 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.15 |
-3.88 |
-51.46 |
2 |
7 |
-1 |
113 |
221.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
501 |
0.52 |
Binding ≤ 10μM
|
|
ATP4A-2-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.50 |
Functional ≤ 10μM
|
|
ATP4B-2-E |
Potassium-transporting ATPase Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
8.62 |
-36.92 |
4 |
6 |
1 |
90 |
253.355 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
0.14 |
7.63 |
-35.87 |
4 |
6 |
1 |
90 |
253.355 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
0.14 |
7.9 |
-14.24 |
3 |
6 |
0 |
89 |
252.347 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
328 |
0.53 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
290 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.06 |
-5.31 |
-19.65 |
4 |
7 |
0 |
118 |
297.745 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.04 |
2.34 |
-13.69 |
3 |
6 |
0 |
98 |
250.283 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.04 |
2.01 |
-53.73 |
2 |
6 |
-1 |
100 |
249.275 |
3 |
↓
|
|
|
|
Analogs
-
37053918
-
-
37053919
-
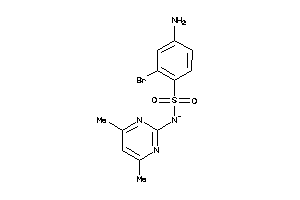
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
3.5 |
-13.79 |
3 |
6 |
0 |
98 |
278.337 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.84 |
3.2 |
-57.09 |
2 |
6 |
-1 |
100 |
277.329 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1700 |
0.50 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1700 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.21 |
-4.95 |
2 |
5 |
0 |
75 |
226.276 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.34 |
-1.29 |
-36.4 |
1 |
5 |
-1 |
82 |
225.268 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1700 |
0.50 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
1700 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.22 |
-4.96 |
2 |
5 |
0 |
75 |
226.276 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.34 |
-1.29 |
-36.41 |
1 |
5 |
-1 |
82 |
225.268 |
4 |
↓
|
|
|
|
Analogs
-
1233255
-
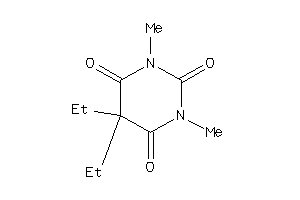
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
1.48 |
-5.14 |
1 |
5 |
0 |
66 |
198.222 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.51 |
-1.08 |
-36.85 |
0 |
5 |
-1 |
73 |
197.214 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.10 |
6.77 |
-3 |
2 |
2 |
0 |
40 |
406.907 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.10 |
8.19 |
-81.93 |
0 |
2 |
-2 |
46 |
404.891 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.10 |
7.44 |
-29.19 |
1 |
2 |
-1 |
43 |
405.899 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
2330 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
3.69 |
-5.63 |
1 |
2 |
0 |
33 |
305.502 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.62 |
4.45 |
-38.91 |
0 |
2 |
-1 |
36 |
304.494 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
8790 |
0.51 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
8790 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
3.69 |
-9.87 |
1 |
4 |
0 |
42 |
261.089 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
0.76 |
3.27 |
-10.81 |
1 |
4 |
0 |
42 |
261.089 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.50 |
2.26 |
-19.27 |
3 |
4 |
0 |
72 |
178.191 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
345 |
0.45 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
312 |
0.46 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
645 |
0.43 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3450 |
0.38 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
91 |
0.49 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
449 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
763 |
0.43 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
134 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2459 |
0.39 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
87 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
345 |
0.45 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
91 |
0.49 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
449 |
0.44 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
87 |
0.49 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
763 |
0.43 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
134 |
0.48 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
7.9 |
0.57 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
312 |
0.46 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
645 |
0.43 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
345 |
0.45 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
91 |
0.49 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
449 |
0.44 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
87 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
763 |
0.43 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
134 |
0.48 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
2459 |
0.39 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
7.9 |
0.57 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
312 |
0.46 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
645 |
0.43 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
3450 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
-3.71 |
-17.79 |
4 |
7 |
0 |
118 |
380.662 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.11 |
-4.11 |
-42.69 |
3 |
7 |
-1 |
120 |
379.654 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
345 |
0.45 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
312 |
0.46 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
645 |
0.43 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3450 |
0.38 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
91 |
0.49 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
449 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
763 |
0.43 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
134 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2459 |
0.39 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
87 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
345 |
0.45 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
91 |
0.49 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
449 |
0.44 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
87 |
0.49 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
763 |
0.43 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
134 |
0.48 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
7.9 |
0.57 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
312 |
0.46 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
645 |
0.43 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
345 |
0.45 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
91 |
0.49 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
449 |
0.44 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
87 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
763 |
0.43 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
134 |
0.48 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
2459 |
0.39 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
7.9 |
0.57 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
312 |
0.46 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
645 |
0.43 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
3450 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.11 |
-3.72 |
-17.55 |
4 |
7 |
0 |
118 |
380.662 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.11 |
-4.23 |
-39.73 |
3 |
7 |
-1 |
120 |
379.654 |
2 |
↓
|
|
|
|
Analogs
-
1530797
-
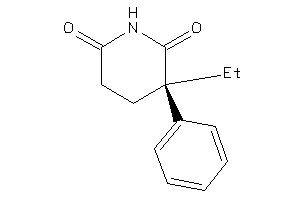
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
4.27 |
-9.63 |
1 |
3 |
0 |
46 |
217.268 |
2 |
↓
|
|
|
|
Analogs
-
1530796
-
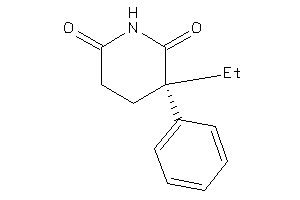
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
-0.06 |
-9.52 |
1 |
3 |
0 |
46 |
217.268 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
7.55 |
-6.8 |
1 |
2 |
0 |
37 |
302.458 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
-4.28 |
-13.32 |
4 |
4 |
0 |
75 |
76.055 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.94 |
-3.07 |
-51.8 |
3 |
4 |
-1 |
78 |
75.047 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.4 |
-10.32 |
3 |
4 |
0 |
68 |
137.142 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.13 |
-0.85 |
-14.04 |
3 |
7 |
0 |
100 |
182.187 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-0.13 |
-0.87 |
-19.56 |
3 |
7 |
0 |
100 |
182.187 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-0.13 |
0.15 |
-14 |
3 |
7 |
0 |
100 |
182.187 |
3 |
↓
|
|
|
|
Analogs
-
1530856
-
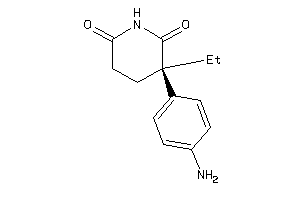
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.15 |
-10.17 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
Analogs
-
1530855
-
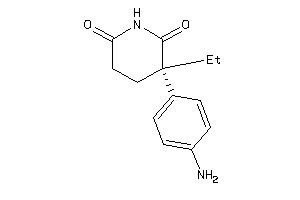
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.19 |
-10.22 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 1μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
0.1 |
0.74 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
5.28 |
-7.85 |
1 |
3 |
0 |
41 |
270.719 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.86 |
6.15 |
-31.26 |
2 |
3 |
1 |
43 |
271.727 |
1 |
↓
|
|
|
|
Analogs
-
643143
-
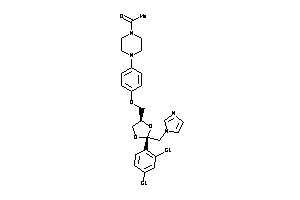
-
643153
-
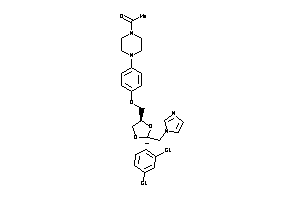
-
3872994
-
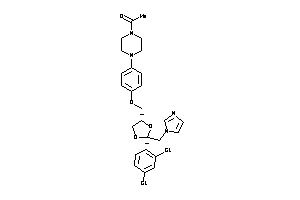
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.26 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
81 |
0.28 |
Binding ≤ 10μM
|
|
CP11A-2-E |
Cytochrome P450 11A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.20 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Binding ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.24 |
Binding ≤ 10μM
|
|
CP21A-1-E |
Cytochrome P450 21 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9010 |
0.20 |
Binding ≤ 10μM
|
|
CP24A-1-E |
Cytochrome P450 24A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
CP3A4-1-E |
Cytochrome P450 3A4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
57 |
0.28 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.28 |
Binding ≤ 10μM
|
|
CP7A1-1-E |
Cytochrome P450 7A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Binding ≤ 10μM
|
|
GNRHR-1-E |
Gonadotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
811 |
0.24 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5720 |
0.20 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.23 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2780 |
0.22 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.21 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.21 |
Functional ≤ 10μM
|
|
CP24A-1-E |
Cytochrome P450 24A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.24 |
ADME/T ≤ 10μM |
|
CP2A2-1-E |
Cytochrome P450 2A2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
543 |
0.24 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
786 |
0.24 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
200 |
0.26 |
Functional ≤ 10μM
|
|
Z50274-1-O |
Aspergillus Flavus (cluster #1 Of 2), Other |
Other |
2800 |
0.22 |
Functional ≤ 10μM
|
|
Z50275-1-O |
Aspergillus Niger (cluster #1 Of 2), Other |
Other |
6000 |
0.20 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
400 |
0.25 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
3200 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.20 |
Functional ≤ 10μM
|
|
Z50436-1-O |
Filobasidiella Neoformans (cluster #1 Of 3), Other |
Other |
20 |
0.30 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.25 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
700 |
0.24 |
Functional ≤ 10μM
|
|
Z50444-1-O |
Candida Parapsilosis (cluster #1 Of 2), Other |
Other |
40 |
0.29 |
Functional ≤ 10μM
|
|
Z50446-1-O |
Candida Tropicalis (cluster #1 Of 2), Other |
Other |
40 |
0.29 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
20 |
0.30 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
2000 |
0.22 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
9800 |
0.19 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
37 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1_BOVIN |
P15150
|
Cytochrome P450 11B1, Bovin |
152 |
0.27 |
Binding ≤ 1μM
|
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
127 |
0.27 |
Binding ≤ 1μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
67 |
0.28 |
Binding ≤ 1μM
|
|
CP17A_RAT |
P11715
|
Cytochrome P450 17A1, Rat |
206 |
0.26 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
26 |
0.29 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
40 |
0.29 |
Binding ≤ 1μM
|
|
CP24A_HUMAN |
Q07973
|
Cytochrome P450 24A1, Human |
312 |
0.25 |
Binding ≤ 1μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
57 |
0.28 |
Binding ≤ 1μM
|
|
CP51A_RAT |
Q64654
|
Cytochrome P450 51, Rat |
119 |
0.27 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
110 |
0.27 |
Binding ≤ 1μM
|
|
CP7A1_RAT |
P18125
|
Cytochrome P450 7A1, Rat |
195 |
0.26 |
Binding ≤ 1μM
|
|
GNRHR_RAT |
P30969
|
Gonadotropin-releasing Hormone Receptor, Rat |
0.221 |
0.38 |
Binding ≤ 1μM
|
|
CP11A_RAT |
P14137
|
Cytochrome P450 11A1, Rat |
1240 |
0.23 |
Binding ≤ 10μM
|
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
127 |
0.27 |
Binding ≤ 10μM
|
|
C11B1_BOVIN |
P15150
|
Cytochrome P450 11B1, Bovin |
152 |
0.27 |
Binding ≤ 10μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
67 |
0.28 |
Binding ≤ 10μM
|
|
CP17A_RAT |
P11715
|
Cytochrome P450 17A1, Rat |
1067 |
0.23 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
2400 |
0.22 |
Binding ≤ 10μM
|
|
CP21A_HUMAN |
P08686
|
Cytochrome P450 21, Human |
9010 |
0.20 |
Binding ≤ 10μM
|
|
CP24A_HUMAN |
Q07973
|
Cytochrome P450 24A1, Human |
312 |
0.25 |
Binding ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
57 |
0.28 |
Binding ≤ 10μM
|
|
CP51A_RAT |
Q64654
|
Cytochrome P450 51, Rat |
119 |
0.27 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
110 |
0.27 |
Binding ≤ 10μM
|
|
CP7A1_RAT |
P18125
|
Cytochrome P450 7A1, Rat |
195 |
0.26 |
Binding ≤ 10μM
|
|
GNRHR_RAT |
P30969
|
Gonadotropin-releasing Hormone Receptor, Rat |
0.221 |
0.38 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1899.98425 |
0.22 |
Binding ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1200 |
0.23 |
Binding ≤ 10μM
|
|
Z50274 |
Z50274
|
Aspergillus Flavus |
2800 |
0.22 |
Functional ≤ 10μM
|
|
Z50416 |
Z50416
|
Aspergillus Fumigatus |
2700 |
0.22 |
Functional ≤ 10μM
|
|
Z50275 |
Z50275
|
Aspergillus Niger |
6000 |
0.20 |
Functional ≤ 10μM
|
|
Z50442 |
Z50442
|
Candida Albicans |
10 |
0.31 |
Functional ≤ 10μM
|
|
Z50443 |
Z50443
|
Candida Glabrata |
700 |
0.24 |
Functional ≤ 10μM
|
|
Z50444 |
Z50444
|
Candida Parapsilosis |
40 |
0.29 |
Functional ≤ 10μM
|
|
Z50446 |
Z50446
|
Candida Tropicalis |
40 |
0.29 |
Functional ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
2780 |
0.22 |
Functional ≤ 10μM
|
|
Z50436 |
Z50436
|
Filobasidiella Neoformans |
100 |
0.27 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
37 |
0.29 |
Functional ≤ 10μM
|
|
Z50408 |
Z50408
|
Issatchenkia Orientalis |
400 |
0.25 |
Functional ≤ 10μM
|
|
Z50459 |
Z50459
|
Leishmania Donovani |
2000 |
0.22 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1000 |
0.23 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
6700 |
0.20 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
3800 |
0.21 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.20 |
Functional ≤ 10μM
|
|
Z102121 |
Z102121
|
Trichophyton Mentagrophytes |
100 |
0.27 |
Functional ≤ 10μM
|
|
Z50452 |
Z50452
|
Trichophyton Rubrum |
10 |
0.31 |
Functional ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
0.3 |
0.37 |
Functional ≤ 10μM
|
|
CP24A_HUMAN |
Q07973
|
Cytochrome P450 24A1, Human |
126 |
0.27 |
ADME/T ≤ 10μM
|
|
CP2A2_RAT |
P15149
|
Cytochrome P450 2A2, Rat |
369 |
0.25 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
11 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
12.55 |
-49.34 |
1 |
8 |
1 |
70 |
532.448 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.69 |
-0.2 |
-10.91 |
0 |
1 |
0 |
17 |
78.136 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80019-3-O |
A-427 (Lung Carcinoma Cells) (cluster #3 Of 4), Other |
Other |
8000 |
0.59 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
8200 |
0.59 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
9000 |
0.59 |
Functional ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
4500 |
0.62 |
Functional ≤ 10μM
|
|
Z80224-13-O |
MCF7 (Breast Carcinoma Cells) (cluster #13 Of 14), Other |
Other |
4500 |
0.62 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
5900 |
0.61 |
Functional ≤ 10μM
|
|
Z80565-2-O |
U-87 MG (Glioblastoma Cells) (cluster #2 Of 2), Other |
Other |
1420 |
0.68 |
Functional ≤ 10μM
|
|
Z80742-3-O |
C6 (Glioma Cells) (cluster #3 Of 3), Other |
Other |
9300 |
0.59 |
Functional ≤ 10μM
|
|
Z80876-1-O |
NCI-H125 (cluster #1 Of 1), Other |
Other |
9000 |
0.59 |
Functional ≤ 10μM
|
|
Z81184-4-O |
LOX IMVI (Melanoma Cells) (cluster #4 Of 5), Other |
Other |
7000 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
7400 |
0.60 |
Functional ≤ 10μM
|
|
Z81281-5-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #5 Of 5), Other |
Other |
5000 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
2.01 |
-5.18 |
1 |
5 |
0 |
62 |
214.052 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
4.1 |
-3.9 |
1 |
5 |
0 |
62 |
233.699 |
4 |
↓
|
|
|
|
Analogs
-
622126
-
-
537809
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
4.81 |
-12.73 |
0 |
6 |
0 |
71 |
352.77 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABPL-3-E |
Fatty Acid-binding Protein, Liver (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9600 |
0.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
6000 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
7.71 |
-6.06 |
0 |
3 |
0 |
36 |
242.702 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4100 |
0.69 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9230 |
0.64 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
6200 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7100 |
0.66 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
0.54 |
-10.06 |
2 |
3 |
0 |
49 |
151.165 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.50 |
Binding ≤ 10μM
|
|
KCNH2-3-E |
HERG (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4898 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.06 |
4.9 |
-11.47 |
0 |
6 |
0 |
62 |
194.194 |
0 |
↓
|
|
|
|
Analogs
-
509440
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
1.04 |
-8.23 |
2 |
4 |
0 |
62 |
286.718 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.02 |
-1.08 |
-50.39 |
1 |
4 |
-1 |
68 |
285.71 |
1 |
↓
|
|
|
|
Analogs
-
575
-
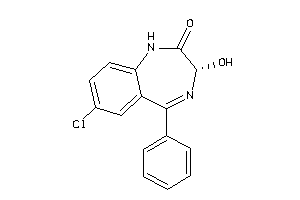
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
2.21 |
-11.02 |
2 |
4 |
0 |
62 |
286.718 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.02 |
0.08 |
-55.63 |
1 |
4 |
-1 |
68 |
285.71 |
1 |
↓
|
|
|
|
Analogs
-
896595
-
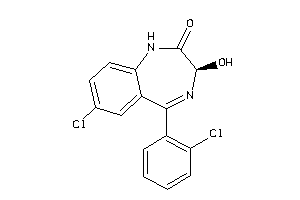
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
1.26 |
-9.16 |
2 |
4 |
0 |
62 |
321.163 |
1 |
↓
|
|
|
|
Analogs
-
431
-
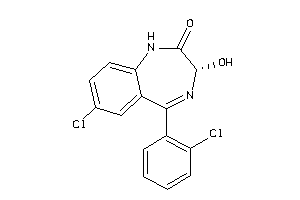
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
-6.06 |
-11.73 |
2 |
4 |
0 |
61 |
321.163 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
8822 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
2.67 |
-11.38 |
6 |
7 |
0 |
130 |
253.269 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.82 |
3.17 |
-25.15 |
7 |
7 |
1 |
131 |
254.277 |
1 |
↓
|
|
|
|
Analogs
-
897283
-
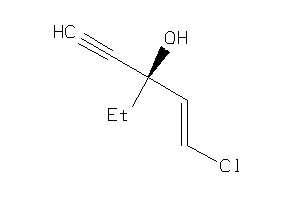
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
0.94 |
-3.12 |
1 |
1 |
0 |
20 |
144.601 |
2 |
↓
|
|
|
|
Analogs
-
267
-
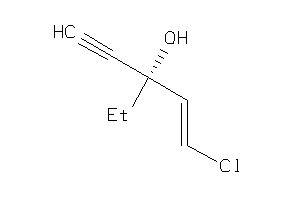
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
0.93 |
-4.22 |
1 |
1 |
0 |
20 |
144.601 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.46 |
15.79 |
-5.38 |
2 |
2 |
0 |
40 |
516.857 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.51 |
-4.55 |
-7.5 |
2 |
4 |
0 |
70 |
123.115 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
4742540
-
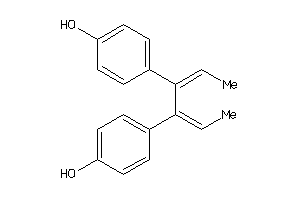
-
38664631
-
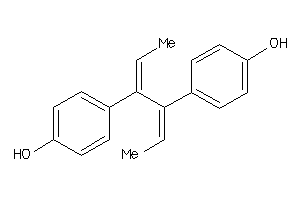
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
5.95 |
-5.56 |
2 |
2 |
0 |
40 |
266.34 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-4-E |
Acetylcholinesterase (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
Binding ≤ 10μM
|
|
EST1-7-E |
Carboxylesterase (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.93 |
Binding ≤ 10μM
|
|
PPGB-1-E |
Lysosomal Protective Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
2.65 |
-7.48 |
0 |
3 |
0 |
35 |
184.147 |
4 |
↓
|
|
|
|
Analogs
-
189374
-
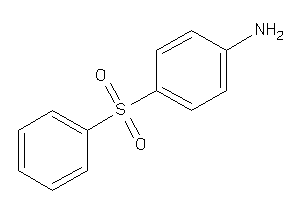
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
1500 |
0.48 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
1500 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
0.99 |
-11.28 |
4 |
4 |
0 |
86 |
248.307 |
2 |
↓
|
|
|
|
Analogs
-
57255
-
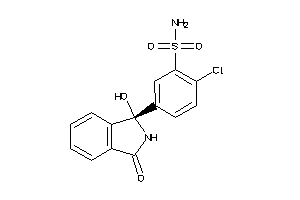
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
348 |
0.41 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4130 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
138 |
0.44 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
196 |
0.43 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
917 |
0.38 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1347 |
0.37 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
23 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
348 |
0.41 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
138 |
0.44 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
196 |
0.43 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
23 |
0.49 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
917 |
0.38 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9 |
0.51 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
2.8 |
0.54 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4.5 |
0.53 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
15 |
0.50 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
348 |
0.41 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
138 |
0.44 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
196 |
0.43 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
23 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
917 |
0.38 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9 |
0.51 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
1347 |
0.37 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
2.8 |
0.54 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4.5 |
0.53 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
4130 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
-2.11 |
-15.19 |
4 |
6 |
0 |
109 |
338.772 |
2 |
↓
|
|
|
|
Analogs
-
20253
-
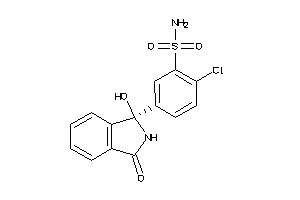
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
348 |
0.41 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4130 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
138 |
0.44 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
196 |
0.43 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
917 |
0.38 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1347 |
0.37 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
23 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
348 |
0.41 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
138 |
0.44 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
196 |
0.43 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
23 |
0.49 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
917 |
0.38 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9 |
0.51 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
2.8 |
0.54 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4.5 |
0.53 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
15 |
0.50 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
348 |
0.41 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
138 |
0.44 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
196 |
0.43 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
23 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
917 |
0.38 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9 |
0.51 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
1347 |
0.37 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
2.8 |
0.54 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4.5 |
0.53 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
4130 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
-2.05 |
-19.26 |
4 |
6 |
0 |
109 |
338.772 |
2 |
↓
|
|