|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-9-E |
Acetylcholinesterase (cluster #9 Of 12), Eukaryotic |
Eukaryotes |
3 |
1.70 |
Binding ≤ 10μM
|
|
SC5A7-1-E |
High Affinity Choline Transporter 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1600 |
1.16 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.24 |
2.42 |
-27.92 |
1 |
2 |
1 |
20 |
104.173 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
7.14 |
-7.63 |
1 |
2 |
0 |
37 |
270.372 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.53 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
|
SHBG-1-E |
Testis-specific Androgen-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Functional ≤ 10μM |
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Functional ≤ 10μM |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Functional ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z80226-1-O |
MCF7-2A (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 1μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 1μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 1μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 1μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 1μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
19.1 |
0.54 |
Binding ≤ 10μM
|
|
ESR1_BOVIN |
P49884
|
Estrogen Receptor Alpha, Bovin |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR1_MOUSE |
P19785
|
Estrogen Receptor Alpha, Mouse |
2.2 |
0.61 |
Binding ≤ 10μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_MOUSE |
O08537
|
Estrogen Receptor Beta, Mouse |
0.5 |
0.65 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.354 |
0.66 |
Binding ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
3.6 |
0.59 |
Binding ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
3.2 |
0.59 |
Binding ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
5.7 |
0.58 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
50 |
0.51 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
0.14 |
0.69 |
Functional ≤ 10μM
|
|
ERR1_HUMAN |
P11474
|
Estrogen-related Receptor Alpha, Human |
5.9 |
0.58 |
Functional ≤ 10μM
|
|
ERR2_HUMAN |
O95718
|
Estrogen-related Receptor Beta, Human |
4.6 |
0.58 |
Functional ≤ 10μM
|
|
GPER_HUMAN |
Q99527
|
G-protein Coupled Estrogen Receptor 1, Human |
0.3 |
0.67 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
0.59 |
0.65 |
Functional ≤ 10μM
|
|
Z81068 |
Z81068
|
Ishikawa (Uterine Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80226 |
Z80226
|
MCF7-2A |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
0.1 |
0.70 |
Functional ≤ 10μM
|
|
ADO_RAT |
Q9Z0U5
|
Aldehyde Oxidase, Rat |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
ADO_HUMAN |
Q06278
|
Aldehyde Oxidase, Human |
80 |
0.50 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
4.8 |
-5.19 |
2 |
2 |
0 |
40 |
272.388 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
4015531
-
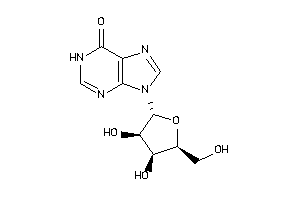
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.82 |
-4.43 |
-23.87 |
4 |
9 |
0 |
133 |
268.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.36 |
-6.52 |
-59.54 |
3 |
9 |
-1 |
137 |
267.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.08 |
-9.7 |
-12.42 |
5 |
7 |
0 |
119 |
221.209 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-2.56 |
-13.08 |
-11.05 |
5 |
7 |
0 |
123 |
221.209 |
2 |
↓
|
|
|
|
Analogs
-
1042045
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.08 |
-9.06 |
-16.14 |
5 |
7 |
0 |
119 |
221.209 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.45 |
2.52 |
-31.62 |
3 |
5 |
1 |
76 |
265.362 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.45 |
2.98 |
-89.37 |
4 |
5 |
2 |
77 |
266.37 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GUAD-1-E |
Guanine Deaminase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1960 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
-0.63 |
-11.05 |
3 |
6 |
0 |
94 |
152.113 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.63 |
-2.01 |
-33.4 |
2 |
6 |
-1 |
97 |
151.105 |
0 |
↓
|
|
|
|
Analogs
-
8637880
-
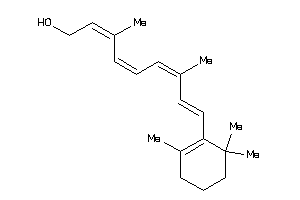
-
12484934
-
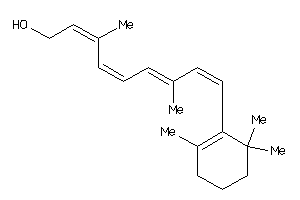
-
12496764
-
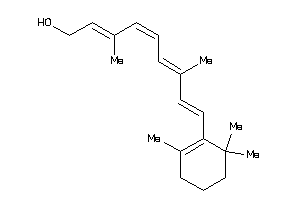
-
12496767
-
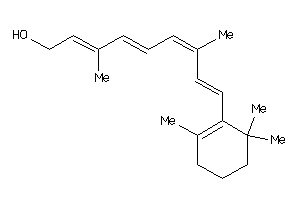
-
12496774
-
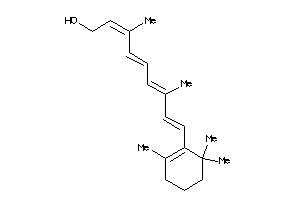
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB-1-E |
Beta-lactoglobulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1-1-E |
Cellular Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2-1-E |
Cellular Retinol-binding Protein II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3-1-E |
Interstitial Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4-1-E |
Plasma Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5-1-E |
Cellular Retinol-binding Protein III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7-1-E |
Cellular Retinol-binding Protein IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 1μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
9.9 |
-4.1 |
1 |
1 |
0 |
20 |
286.459 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2620 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
2620 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.33 |
-5.82 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3947571
-
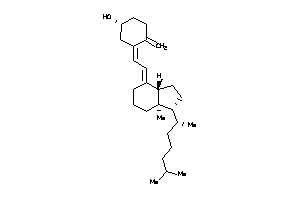
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-1-E |
Dual Specificity Phosphatase Cdc25A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.53 |
-2.24 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
-
3947571
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPIP1-1-E |
Dual Specificity Phosphatase Cdc25A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.59 |
-2.14 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
1718319
-
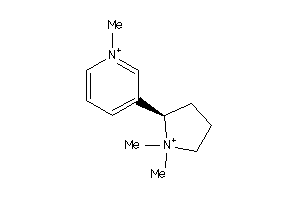
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.16 |
5.88 |
-27.75 |
0 |
2 |
1 |
7 |
177.271 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.05 |
5.69 |
-35.01 |
0 |
2 |
1 |
16 |
161.228 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.05 |
5.98 |
-87.61 |
1 |
2 |
2 |
17 |
162.236 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
3100 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
8900 |
0.64 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9200 |
0.64 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
48 |
0.93 |
Binding ≤ 10μM
|
|
CAH2-15-E |
Carbonic Anhydrase II (cluster #15 Of 15), Eukaryotic |
Eukaryotes |
9200 |
0.64 |
Binding ≤ 10μM
|
|
CAH3-6-E |
Carbonic Anhydrase III (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5600 |
0.67 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
6800 |
0.66 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
9400 |
0.64 |
Binding ≤ 10μM
|
|
CAH5B-6-E |
Carbonic Anhydrase VB (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
5200 |
0.67 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
2000 |
0.73 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
3300 |
0.70 |
Binding ≤ 10μM
|
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
48 |
0.93 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
3100 |
0.70 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
9200 |
0.64 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
5600 |
0.67 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
6800 |
0.66 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
9100 |
0.64 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
9400 |
0.64 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
5200 |
0.67 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
2000 |
0.73 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
3300 |
0.70 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
8900 |
0.64 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
9200 |
0.64 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
48 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
5.99 |
-11.08 |
0 |
2 |
0 |
30 |
146.145 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1640 |
0.51 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1640 |
0.51 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1640 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
1.46 |
-14.44 |
3 |
4 |
0 |
65 |
218.256 |
3 |
↓
|
|
|
|
Analogs
-
3830982
-
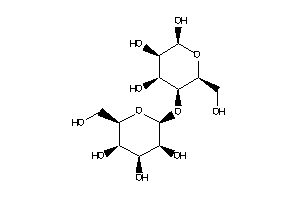
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.25 |
-23.42 |
-16 |
11 |
16 |
0 |
269 |
504.438 |
7 |
↓
|
|
|
|
Analogs
-
3830982
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.25 |
-23.55 |
-15.09 |
11 |
16 |
0 |
269 |
504.438 |
7 |
↓
|
|
|
|
Analogs
-
2379161
-
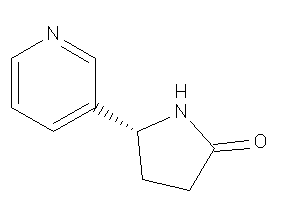
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.17 |
-1.82 |
-10.74 |
1 |
3 |
0 |
41 |
162.192 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.17 |
-1.71 |
-39.86 |
2 |
3 |
1 |
43 |
163.2 |
1 |
↓
|
|
|
|
Analogs
-
3869576
-
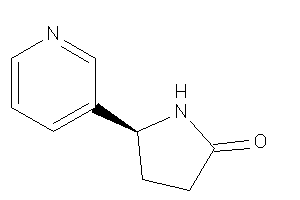
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.17 |
-1.82 |
-10.73 |
1 |
3 |
0 |
41 |
162.192 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.17 |
-1.71 |
-39.86 |
2 |
3 |
1 |
43 |
163.2 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
4.03 |
-3.28 |
0 |
3 |
0 |
39 |
117.148 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
1.06 |
-11.82 |
1 |
4 |
0 |
53 |
192.218 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.50 |
-2.49 |
-40.85 |
2 |
4 |
1 |
54 |
193.226 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.50 |
1.6 |
-42.29 |
2 |
4 |
1 |
55 |
193.226 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
2.14 |
-6.64 |
1 |
2 |
0 |
29 |
68.079 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
2.68 |
-31.31 |
2 |
2 |
1 |
30 |
69.087 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
3.75 |
-4.16 |
2 |
2 |
0 |
30 |
69.087 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.50 |
Binding ≤ 10μM
|
|
KCNH2-3-E |
HERG (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4898 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.06 |
4.9 |
-11.47 |
0 |
6 |
0 |
62 |
194.194 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4100 |
0.69 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9230 |
0.64 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
6200 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7100 |
0.66 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
0.54 |
-10.06 |
2 |
3 |
0 |
49 |
151.165 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
3.18 |
-11.02 |
1 |
6 |
0 |
73 |
180.167 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-3.42 |
-5.18 |
2 |
2 |
0 |
40 |
76.095 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-3.42 |
-5.18 |
2 |
2 |
0 |
40 |
76.095 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.02 |
1.1 |
-11.24 |
2 |
6 |
0 |
84 |
166.14 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.54 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.54 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.54 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.85 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
9000 |
0.54 |
Binding ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
6520 |
0.56 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
0.6 |
0.99 |
Binding ≤ 1μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.99 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
14 |
0.85 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
0.6 |
0.99 |
Binding ≤ 10μM
|
|
AA1R_BOVIN |
P28190
|
Adenosine A1 Receptor, Bovin |
2430 |
0.60 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
1310 |
0.63 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.99 |
Binding ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
1310 |
0.63 |
Binding ≤ 10μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
2700 |
0.60 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
14 |
0.85 |
Binding ≤ 10μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
150 |
0.73 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
9070 |
0.54 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
10000 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
3.24 |
-11.33 |
1 |
6 |
0 |
73 |
180.167 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
6 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.39 |
Binding ≤ 10μM
|
|
KCNA3-1-E |
Voltage-gated Potassium Channel Subunit Kv1.3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.31 |
Binding ≤ 10μM
|
|
KCNN4-1-E |
Intermediate Conductance Calcium-activated Potassium Channel Protein 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.40 |
Binding ≤ 10μM
|
|
MDHM-1-E |
Malate Dehydrogenase, Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.29 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.41 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
103 |
0.39 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
4790 |
0.30 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6540 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
60 |
0.40 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-8-O |
A549 (Lung Carcinoma Cells) (cluster #8 Of 11), Other |
Other |
5100 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-2-O |
NIH3T3 (Fibroblasts) (cluster #2 Of 4), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
3000 |
0.31 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q96W81_ASPFM |
Q96W81
|
14-alpha Sterol Demethylase, Aspfm |
103 |
0.39 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
24 |
0.43 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
81.5 |
0.40 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1.8 |
0.49 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
130 |
0.39 |
Binding ≤ 1μM
|
|
KCNN4_HUMAN |
O15554
|
Intermediate Conductance Calcium-activated Potassium Channel Protein 4, Human |
360 |
0.36 |
Binding ≤ 1μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 1μM
|
|
MDHM_PIG |
P00346
|
Malate Dehydrogenase Mitochondrial, Pig |
2 |
0.49 |
Binding ≤ 1μM
|
|
Q96W81_ASPFM |
Q96W81
|
14-alpha Sterol Demethylase, Aspfm |
103 |
0.39 |
Binding ≤ 10μM
|
|
Q4WNT5_ASPFU |
Q4WNT5
|
14-alpha Sterol Demethylase Cyp51A, Aspfu |
4790 |
0.30 |
Binding ≤ 10μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
24 |
0.43 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
81.5 |
0.40 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1.8 |
0.49 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
130 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
3019.95172 |
0.31 |
Binding ≤ 10μM
|
|
KCNN4_HUMAN |
O15554
|
Intermediate Conductance Calcium-activated Potassium Channel Protein 4, Human |
360 |
0.36 |
Binding ≤ 10μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 10μM
|
|
MDHM_PIG |
P00346
|
Malate Dehydrogenase Mitochondrial, Pig |
2 |
0.49 |
Binding ≤ 10μM
|
|
KCNA3_HUMAN |
P22001
|
Voltage-gated Potassium Channel Subunit Kv1.3, Human |
6000 |
0.29 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
5100 |
0.30 |
Functional ≤ 10μM
|
|
Z50380 |
Z50380
|
Mycobacterium Smegmatis |
6540 |
0.29 |
Functional ≤ 10μM
|
|
Z80951 |
Z80951
|
NIH3T3 (Fibroblasts) |
630 |
0.35 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1300 |
0.33 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
3500 |
0.31 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
4800 |
0.30 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
250 |
0.37 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
20 |
0.43 |
ADME/T ≤ 10μM
|
|
Z80936 |
Z80936
|
HEK293 (Embryonic Kidney Fibroblasts) |
3000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
13.43 |
-5.78 |
0 |
2 |
0 |
18 |
344.845 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
13.99 |
-28.18 |
1 |
2 |
1 |
19 |
345.853 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
328 |
0.53 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
290 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.06 |
-5.31 |
-19.65 |
4 |
7 |
0 |
118 |
297.745 |
1 |
↓
|
|
|
|
Analogs
-
2944385
-
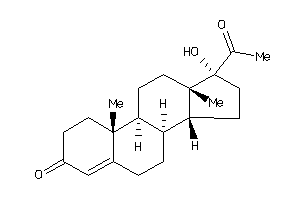
-
3651055
-
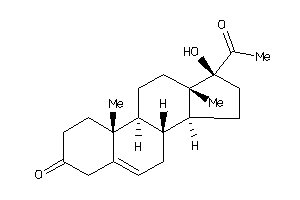
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
7.96 |
-9.5 |
1 |
3 |
0 |
54 |
344.495 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-15-O |
Plasmodium Falciparum (cluster #15 Of 22), Other |
Other |
8822 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
2.67 |
-11.38 |
6 |
7 |
0 |
130 |
253.269 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.82 |
3.17 |
-25.15 |
7 |
7 |
1 |
131 |
254.277 |
1 |
↓
|
|