|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.04 |
4.06 |
-54.8 |
1 |
4 |
-1 |
69 |
137.118 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.28 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.22 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.31 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
5.67 |
-58.98 |
0 |
8 |
-1 |
105 |
453.475 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
4.94 |
6.79 |
-50.07 |
1 |
8 |
-1 |
105 |
453.475 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.94 |
7.33 |
-15.42 |
2 |
8 |
0 |
102 |
454.483 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B-2-E |
Heat Shock Protein HSP 90-beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1560 |
0.35 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90B_HUMAN |
P08238
|
Heat Shock Protein HSP 90-beta, Human |
1560 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
9.15 |
-6.77 |
2 |
5 |
0 |
77 |
344.205 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
5.11 |
7.77 |
-6.42 |
2 |
5 |
0 |
77 |
344.205 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
8.65 |
-3.67 |
0 |
0 |
0 |
0 |
356.883 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104305-1-O |
Tumour Suppressor P53/oncoprotein Mdm2 (cluster #1 Of 1), Other |
Other |
1230 |
0.27 |
Binding ≤ 10μM
|
|
Z104305-1-O |
Tumour Suppressor P53/oncoprotein Mdm2 (cluster #1 Of 1), Other |
Other |
3880 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104305 |
Z104305
|
Tumour Suppressor P53/oncoprotein Mdm2 |
1230 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.27 |
14.37 |
-13.95 |
0 |
5 |
0 |
52 |
536.227 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.27 |
14.89 |
-12.25 |
1 |
5 |
0 |
54 |
537.235 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.95 |
1.8 |
-50.85 |
3 |
5 |
-1 |
80 |
166.189 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.12 |
2.33 |
-39.26 |
3 |
5 |
-1 |
81 |
166.189 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.12 |
2.34 |
-38.24 |
3 |
5 |
-1 |
81 |
166.189 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
9.26 |
-50.16 |
3 |
7 |
1 |
80 |
383.501 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.41 |
5.94 |
-12.37 |
1 |
7 |
0 |
75 |
382.493 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
7.97 |
-10.28 |
2 |
7 |
0 |
75 |
382.493 |
4 |
↓
|
|
|
|
Analogs
-
3650249
-
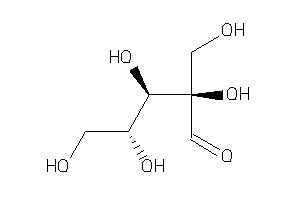
-
8552145
-
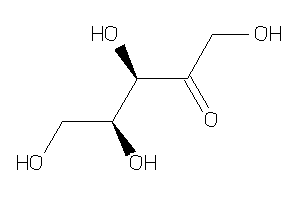
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.22 |
-7.48 |
-11.99 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-2.22 |
-7.36 |
-11.73 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-2.22 |
-5.76 |
-11.37 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.23 |
17.08 |
-50.99 |
2 |
8 |
1 |
80 |
559.119 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.23 |
14.72 |
-13.89 |
1 |
8 |
0 |
79 |
558.111 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.23 |
17.84 |
-127.12 |
3 |
8 |
2 |
82 |
560.127 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.56 |
-2.91 |
-56.93 |
4 |
4 |
0 |
80 |
130.147 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.56 |
-2.19 |
-41.97 |
3 |
4 |
-1 |
78 |
129.139 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.56 |
-1.77 |
-56.67 |
4 |
4 |
0 |
83 |
130.147 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.25 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.26 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
108 |
0.24 |
Binding ≤ 10μM
|
|
FGFR4-1-E |
Fibroblast Growth Factor Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
610 |
0.22 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.27 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
195 |
0.23 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.25 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
156 |
0.24 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.26 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.29 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.29 |
Binding ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.28 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
38 |
0.26 |
Binding ≤ 1μM
|
|
FGFR2_HUMAN |
P21802
|
Fibroblast Growth Factor Receptor 2, Human |
37 |
0.26 |
Binding ≤ 1μM
|
|
FGFR3_HUMAN |
P22607
|
Fibroblast Growth Factor Receptor 3, Human |
108 |
0.24 |
Binding ≤ 1μM
|
|
FGFR4_HUMAN |
P22455
|
Fibroblast Growth Factor Receptor 4, Human |
610 |
0.22 |
Binding ≤ 1μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
18 |
0.27 |
Binding ≤ 1μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
65 |
0.25 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
16 |
0.27 |
Binding ≤ 1μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
195 |
0.23 |
Binding ≤ 1μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
26 |
0.27 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
156 |
0.24 |
Binding ≤ 1μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
104 |
0.24 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
21 |
0.27 |
Binding ≤ 1μM
|
|
VGFR2_MOUSE |
P35918
|
Vascular Endothelial Growth Factor Receptor 2, Mouse |
13 |
0.28 |
Binding ≤ 1μM
|
|
VGFR3_HUMAN |
P35916
|
Vascular Endothelial Growth Factor Receptor 3, Human |
13 |
0.28 |
Binding ≤ 1μM
|
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
38 |
0.26 |
Binding ≤ 10μM
|
|
FGFR2_HUMAN |
P21802
|
Fibroblast Growth Factor Receptor 2, Human |
37 |
0.26 |
Binding ≤ 10μM
|
|
FGFR3_HUMAN |
P22607
|
Fibroblast Growth Factor Receptor 3, Human |
108 |
0.24 |
Binding ≤ 10μM
|
|
FGFR4_HUMAN |
P22455
|
Fibroblast Growth Factor Receptor 4, Human |
610 |
0.22 |
Binding ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
18 |
0.27 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
65 |
0.25 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
16 |
0.27 |
Binding ≤ 10μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
195 |
0.23 |
Binding ≤ 10μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
26 |
0.27 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
156 |
0.24 |
Binding ≤ 10μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
104 |
0.24 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
21 |
0.27 |
Binding ≤ 10μM
|
|
VGFR2_MOUSE |
P35918
|
Vascular Endothelial Growth Factor Receptor 2, Mouse |
13 |
0.28 |
Binding ≤ 10μM
|
|
VGFR3_HUMAN |
P35916
|
Vascular Endothelial Growth Factor Receptor 3, Human |
13 |
0.28 |
Binding ≤ 10μM
|
|
Z81057 |
Z81057
|
HUVEC (Umbilical Vein Endothelial Cells) |
10 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
11.6 |
-47.6 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
4.16 |
11.94 |
-48.76 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
4.62 |
8.82 |
-50.42 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT2-2-E |
Serine/threonine-protein Kinase AKT2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
6.63 |
-56.58 |
5 |
7 |
1 |
102 |
385.879 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.83 |
6.36 |
-11.34 |
4 |
7 |
0 |
100 |
384.871 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.34 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
100 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
7.28 |
-44.08 |
1 |
4 |
-1 |
73 |
309.341 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.22 |
6.68 |
-18.6 |
2 |
4 |
0 |
71 |
310.349 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.22 |
6.07 |
-17.85 |
2 |
4 |
0 |
71 |
310.349 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
VDR-1-E |
Vitamin D Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
VDR-1-E |
Vitamin D Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
52 |
0.34 |
Functional ≤ 10μM
|
|
Z80437-1-O |
RWLeu4 (cluster #1 Of 1), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM |
|
Z80437-1-O |
RWLeu4 (cluster #1 Of 1), Other |
Other |
6 |
0.38 |
Functional ≤ 10μM |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
10 |
0.37 |
Functional ≤ 10μM |
|
Z80566-2-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #2 Of 2), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM |
|
Z81117-2-O |
Keratinocytes (Keratinocytes) (cluster #2 Of 2), Other |
Other |
27 |
0.35 |
Functional ≤ 10μM
|
|
Z81298-1-O |
PE Cell Line (cluster #1 Of 1), Other |
Other |
100 |
0.33 |
Functional ≤ 10μM |
|
Z81338-2-O |
T-cells (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
6.98 |
-5.55 |
3 |
3 |
0 |
61 |
416.646 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
8618002
-
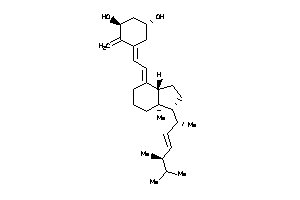
-
26522802
-
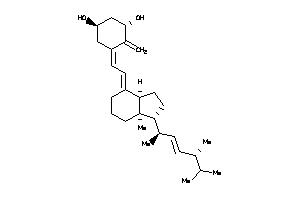
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.51 |
10.74 |
-3.74 |
2 |
2 |
0 |
40 |
412.658 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GUAD-1-E |
Guanine Deaminase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4340 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.99 |
-5.59 |
-30.42 |
3 |
7 |
-1 |
117 |
167.104 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.44 |
-3.48 |
-11.55 |
4 |
7 |
0 |
114 |
168.112 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
5298929
-
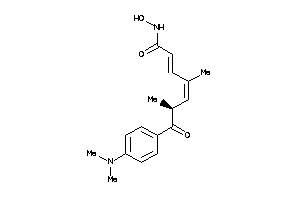
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
700 |
0.39 |
Binding ≤ 10μM
|
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDA10-3-E |
Histone Deacetylase 10 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.33 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC4-3-E |
Histone Deacetylase 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC5-3-E |
Histone Deacetylase 5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
81 |
0.45 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC8-2-E |
Histone Deacetylase 8 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.38 |
Binding ≤ 10μM |
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.39 |
Binding ≤ 10μM
|
|
NCOR2-2-E |
Nuclear Receptor Corepressor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8600 |
0.32 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Q9ZTP8-1-E |
Histone Deacetylase HD1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SF3B3-1-E |
Splicing Factor 3B Subunit 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
611 |
0.40 |
Binding ≤ 10μM
|
|
TF-1-E |
Coagulation Factor III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z103359-1-O |
NCI-H661 (cluster #1 Of 1), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10 |
0.51 |
Functional ≤ 10μM
|
|
Z80123-1-O |
DO4 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80277-1-O |
MM96L (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.48 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
280 |
0.42 |
Functional ≤ 10μM
|
|
Z80513-1-O |
SQ20B (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80525-1-O |
SW48 (cluster #1 Of 2), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z80725-1-O |
C180-13S (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z80857-2-O |
Friend Leukemia Cell Line (cluster #2 Of 2), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z80957-1-O |
HMEC (Microvascular Endothelial Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.37 |
Functional ≤ 10μM
|
|
Z81070-1-O |
JAM Cell Line (cluster #1 Of 1), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z81199-1-O |
NCI-H446 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z80332-1-O |
NFF (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TF_HUMAN |
P13726
|
Coagulation Factor III, Human |
20 |
0.49 |
Binding ≤ 1μM
|
|
Q9XYC7_PLAFA |
Q9XYC7
|
Histone Deacetylase, Plafa |
0.6 |
0.59 |
Binding ≤ 1μM
|
|
HDAC1_RAT |
Q4QQW4
|
Histone Deacetylase 1, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
0.15 |
0.63 |
Binding ≤ 1μM
|
|
HDAC1_MOUSE |
O09106
|
Histone Deacetylase 1, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDA11_MOUSE |
Q91WA3
|
Histone Deacetylase 11, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
0.4 |
0.60 |
Binding ≤ 1μM
|
|
HDAC2_MOUSE |
P70288
|
Histone Deacetylase 2, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
B1WBY8_RAT |
B1WBY8
|
Histone Deacetylase 2, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC3_MOUSE |
O88895
|
Histone Deacetylase 3, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
0.26 |
0.61 |
Binding ≤ 1μM
|
|
HDAC3_RAT |
Q6P6W3
|
Histone Deacetylase 3, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
0.8 |
0.58 |
Binding ≤ 1μM
|
|
HDAC4_RAT |
Q99P99
|
Histone Deacetylase 4, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
Q5RJZ2_RAT |
Q5RJZ2
|
Histone Deacetylase 5, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
0.8 |
0.58 |
Binding ≤ 1μM
|
|
HDAC5_MOUSE |
Q9Z2V6
|
Histone Deacetylase 5, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC6_MOUSE |
Q9Z2V5
|
Histone Deacetylase 6, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
0.13 |
0.63 |
Binding ≤ 1μM
|
|
Q99P97_RAT |
Q99P97
|
Histone Deacetylase 6, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
0.8 |
0.58 |
Binding ≤ 1μM
|
|
HDAC7_MOUSE |
Q8C2B3
|
Histone Deacetylase 7, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC7_RAT |
Q99P96
|
Histone Deacetylase 7, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
0.4 |
0.60 |
Binding ≤ 1μM
|
|
HDAC8_MOUSE |
Q8VH37
|
Histone Deacetylase 8, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
0.8 |
0.58 |
Binding ≤ 1μM
|
|
HDAC9_MOUSE |
Q99N13
|
Histone Deacetylase 9, Mouse |
1.44 |
0.56 |
Binding ≤ 1μM
|
|
Q9ZTP8_MAIZE |
Q9ZTP8
|
Histone Deacetylase HD1B, Maize |
0.4 |
0.60 |
Binding ≤ 1μM
|
|
Q94F81_MAIZE |
Q94F81
|
Histone Deacetylase HD2, Maize |
3 |
0.54 |
Binding ≤ 1μM
|
|
HDAH_ALCSD |
Q70I53
|
Histone Deacetylase-like Amidohydrolase, Alcsd |
700 |
0.39 |
Binding ≤ 1μM
|
|
NCOR2_HUMAN |
Q9Y618
|
Nuclear Receptor Corepressor 2, Human |
0.26 |
0.61 |
Binding ≤ 1μM
|
|
SF3B3_HUMAN |
Q15393
|
Splicing Factor 3B Subunit 3, Human |
577.6 |
0.40 |
Binding ≤ 1μM
|
|
TF_HUMAN |
P13726
|
Coagulation Factor III, Human |
20 |
0.49 |
Binding ≤ 10μM
|
|
Q9XYC7_PLAFA |
Q9XYC7
|
Histone Deacetylase, Plafa |
0.6 |
0.59 |
Binding ≤ 10μM
|
|
HDAC1_MOUSE |
O09106
|
Histone Deacetylase 1, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC1_RAT |
Q4QQW4
|
Histone Deacetylase 1, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
0.15 |
0.63 |
Binding ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDA11_MOUSE |
Q91WA3
|
Histone Deacetylase 11, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
B1WBY8_RAT |
B1WBY8
|
Histone Deacetylase 2, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
0.4 |
0.60 |
Binding ≤ 10μM
|
|
HDAC2_MOUSE |
P70288
|
Histone Deacetylase 2, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC3_RAT |
Q6P6W3
|
Histone Deacetylase 3, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC3_MOUSE |
O88895
|
Histone Deacetylase 3, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
0.26 |
0.61 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
0.8 |
0.58 |
Binding ≤ 10μM
|
|
HDAC4_RAT |
Q99P99
|
Histone Deacetylase 4, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC5_MOUSE |
Q9Z2V6
|
Histone Deacetylase 5, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
Q5RJZ2_RAT |
Q5RJZ2
|
Histone Deacetylase 5, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
0.8 |
0.58 |
Binding ≤ 10μM
|
|
HDAC6_MOUSE |
Q9Z2V5
|
Histone Deacetylase 6, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
0.13 |
0.63 |
Binding ≤ 10μM
|
|
Q99P97_RAT |
Q99P97
|
Histone Deacetylase 6, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC7_RAT |
Q99P96
|
Histone Deacetylase 7, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
0.8 |
0.58 |
Binding ≤ 10μM
|
|
HDAC7_MOUSE |
Q8C2B3
|
Histone Deacetylase 7, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
0.4 |
0.60 |
Binding ≤ 10μM
|
|
HDAC8_MOUSE |
Q8VH37
|
Histone Deacetylase 8, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC9_MOUSE |
Q99N13
|
Histone Deacetylase 9, Mouse |
1.44 |
0.56 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
0.8 |
0.58 |
Binding ≤ 10μM
|
|
Q9ZTP8_MAIZE |
Q9ZTP8
|
Histone Deacetylase HD1B, Maize |
0.4 |
0.60 |
Binding ≤ 10μM
|
|
Q94F81_MAIZE |
Q94F81
|
Histone Deacetylase HD2, Maize |
3 |
0.54 |
Binding ≤ 10μM
|
|
HDAH_ALCSD |
Q70I53
|
Histone Deacetylase-like Amidohydrolase, Alcsd |
1200 |
0.38 |
Binding ≤ 10μM
|
|
NCOR2_HUMAN |
Q9Y618
|
Nuclear Receptor Corepressor 2, Human |
0.26 |
0.61 |
Binding ≤ 10μM
|
|
SF3B3_HUMAN |
Q15393
|
Splicing Factor 3B Subunit 3, Human |
577.6 |
0.40 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
160 |
0.43 |
Functional ≤ 10μM
|
|
Z80725 |
Z80725
|
C180-13S (Ovarian Carcinoma Cells) |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z80123 |
Z80123
|
DO4 (Melanoma Cells) |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z80125 |
Z80125
|
DU-145 (Prostate Carcinoma) |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80857 |
Z80857
|
Friend Leukemia Cell Line |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
1.3 |
0.57 |
Functional ≤ 10μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80957 |
Z80957
|
HMEC (Microvascular Endothelial Cells) |
1600 |
0.37 |
Functional ≤ 10μM
|
|
Z81070 |
Z81070
|
JAM Cell Line |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
3500 |
0.35 |
Functional ≤ 10μM
|
|
Z80269 |
Z80269
|
MKN-45 (Gastric Adenocarcinoma Cells) |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80277 |
Z80277
|
MM96L (Melanoma Cells) |
30 |
0.48 |
Functional ≤ 10μM
|
|
Z80877 |
Z80877
|
NCI-H1299 (Non-small Cell Lung Carcinoma) |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z81199 |
Z81199
|
NCI-H446 (Small Cell Lung Carcinoma Cells) |
1500 |
0.37 |
Functional ≤ 10μM
|
|
Z103359 |
Z103359
|
NCI-H661 |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10 |
0.51 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z80485 |
Z80485
|
SK-MEL-28 (Melanoma Cells) |
280 |
0.42 |
Functional ≤ 10μM
|
|
Z80513 |
Z80513
|
SQ20B |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80525 |
Z80525
|
SW48 |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80532 |
Z80532
|
T-24 (Bladder Carcinoma Cells) |
150 |
0.43 |
Functional ≤ 10μM
|
|
Z80332 |
Z80332
|
NFF (Fibroblast Cells) |
200 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
6.23 |
-20.9 |
2 |
5 |
0 |
70 |
302.374 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
2.68 |
6.23 |
-20.14 |
2 |
5 |
0 |
70 |
302.374 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.68 |
7.44 |
-64.15 |
1 |
5 |
-1 |
72 |
301.366 |
6 |
↓
|
|
|
|
Analogs
-
22172658
-
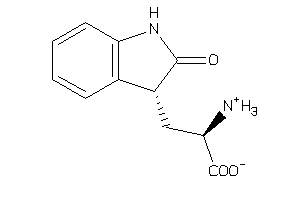
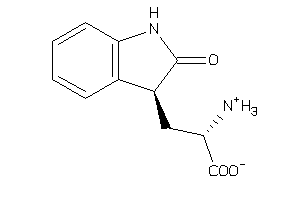
-
1590598
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.18 |
2.18 |
-38.96 |
4 |
5 |
0 |
97 |
220.228 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-1.18 |
0.4 |
-35.02 |
5 |
5 |
0 |
104 |
220.228 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.18 |
0.08 |
-49 |
4 |
5 |
-1 |
102 |
219.22 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
8215517
-
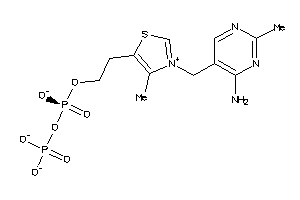
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
0.47 |
-43.06 |
1 |
4 |
-1 |
64 |
152.11 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.19 |
1.59 |
-133.32 |
0 |
4 |
-2 |
66 |
151.102 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
1.9 |
-67.53 |
1 |
4 |
-1 |
68 |
152.11 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
11.28 |
-11.67 |
0 |
6 |
0 |
71 |
380.481 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B2CL1-1-E |
Apoptosis Regulator Bcl-X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.50 |
Binding ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
2100 |
0.50 |
Binding ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
6500 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
-1.18 |
-18.01 |
4 |
5 |
0 |
98 |
220.18 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.69 |
-1.6 |
-19.3 |
4 |
5 |
0 |
98 |
220.18 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.69 |
-0.74 |
-54.35 |
3 |
5 |
-1 |
101 |
219.172 |
0 |
↓
|
|
|
|
Analogs
-
11592545
-
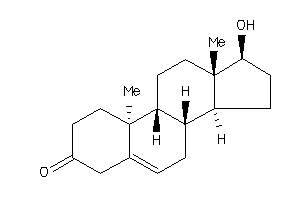
-
11592546
-
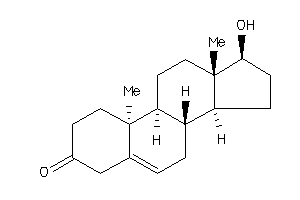
-
103270
-
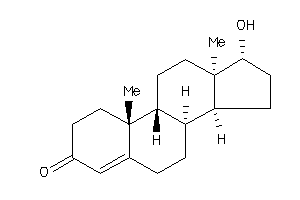
-
490793
-
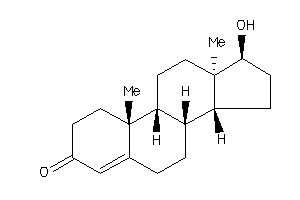
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 10μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 10μM
|
|
ERG2_YEAST |
P32352
|
C-8 Sterol Isomerase, Yeast |
7760 |
0.34 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
1200 |
0.39 |
Binding ≤ 10μM
|
|
S5A1_RAT |
P24008
|
Steroid 5-alpha-reductase 1, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
S5A2_RAT |
P31214
|
Steroid 5-alpha-reductase 2, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 10μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
3.2 |
0.57 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
10000 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.07 |
-7.05 |
1 |
2 |
0 |
37 |
288.431 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.99 |
5.16 |
-4.08 |
2 |
2 |
0 |
40 |
288.431 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
-0.69 |
-0.82 |
-56.84 |
7 |
10 |
0 |
186 |
444.44 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-2-E |
Nitric-oxide Synthase, Brain (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
60 |
0.78 |
Binding ≤ 10μM
|
|
NOS2-7-E |
Nitric Oxide Synthase, Inducible (cluster #7 Of 9), Eukaryotic |
Eukaryotes |
300 |
0.70 |
Binding ≤ 10μM
|
|
NOS3-1-E |
Nitric Oxide Synthase, Endothelial (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2_HUMAN |
P35228
|
Nitric Oxide Synthase, Inducible, Human |
300 |
0.70 |
Binding ≤ 1μM
|
|
NOS1_RAT |
P29476
|
Nitric-oxide Synthase, Brain, Rat |
60 |
0.78 |
Binding ≤ 1μM
|
|
NOS1_HUMAN |
P29475
|
Nitric-oxide Synthase, Brain, Human |
60 |
0.78 |
Binding ≤ 1μM
|
|
NOS3_HUMAN |
P29474
|
Nitric-oxide Synthase, Endothelial, Human |
400 |
0.69 |
Binding ≤ 1μM
|
|
NOS2_HUMAN |
P35228
|
Nitric Oxide Synthase, Inducible, Human |
300 |
0.70 |
Binding ≤ 10μM
|
|
NOS1_HUMAN |
P29475
|
Nitric-oxide Synthase, Brain, Human |
60 |
0.78 |
Binding ≤ 10μM
|
|
NOS1_RAT |
P29476
|
Nitric-oxide Synthase, Brain, Rat |
60 |
0.78 |
Binding ≤ 10μM
|
|
NOS3_HUMAN |
P29474
|
Nitric-oxide Synthase, Endothelial, Human |
400 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.52 |
1.26 |
-72.12 |
6 |
5 |
1 |
105 |
206.291 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.65 |
0.87 |
-52.03 |
5 |
5 |
0 |
106 |
205.283 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.52 |
0.93 |
-52.68 |
5 |
5 |
0 |
104 |
205.283 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DCAM-1-E |
S-adenosylmethionine Decarboxylase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.28 |
Binding ≤ 10μM |
|
PNMT-1-E |
Phenylethanolamine N-methyltransferase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
12402861
-
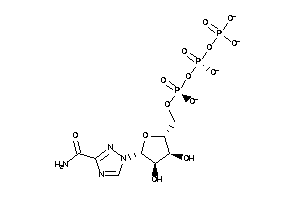
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
12 |
-18.58 |
0 |
3 |
1 |
19 |
343.45 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
5.79 |
-15.08 |
1 |
6 |
0 |
93 |
362.432 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
3.78 |
5.7 |
-14.92 |
1 |
6 |
0 |
93 |
362.432 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
3.79 |
-60.86 |
0 |
6 |
-1 |
96 |
361.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.81 |
0.67 |
-15.35 |
3 |
11 |
0 |
152 |
444.423 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.81 |
1.61 |
-55.71 |
2 |
11 |
-1 |
155 |
443.415 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.74 |
-3.29 |
-50.91 |
2 |
11 |
-1 |
159 |
443.415 |
6 |
↓
|
|