|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
3940 |
0.63 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
3330 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
4.49 |
-53.36 |
0 |
3 |
-1 |
43 |
164.184 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.39 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
3000 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
7.56 |
-45.01 |
1 |
4 |
-1 |
73 |
309.341 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.22 |
6.96 |
-17.09 |
2 |
4 |
0 |
71 |
310.349 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.22 |
6.34 |
-14.56 |
2 |
4 |
0 |
71 |
310.349 |
4 |
↓
|
|
|
|
Analogs
-
3188038
-
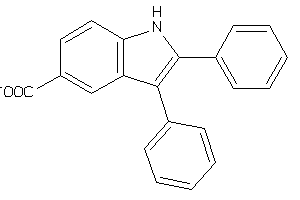
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
12.33 |
-52.43 |
1 |
3 |
-1 |
56 |
312.348 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BIRC5-1-E |
Baculoviral IAP Repeat-containing Protein 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5700 |
0.32 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
5.32 |
-48.47 |
1 |
4 |
-1 |
80 |
366.194 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.66 |
6.09 |
-120.19 |
0 |
4 |
-2 |
83 |
365.186 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.20 |
7.57 |
-36.94 |
1 |
4 |
-1 |
80 |
366.194 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
1.15 |
-156.52 |
0 |
5 |
-2 |
93 |
306.273 |
4 |
↓
|
|
|
|
Analogs
-
35902488
-
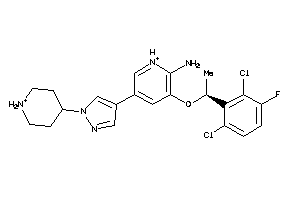
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1159 |
0.28 |
Binding ≤ 10μM |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM |
|
INSR-1-E |
Insulin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.29 |
Binding ≤ 10μM |
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2741 |
0.26 |
Binding ≤ 10μM |
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM |
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.36 |
Binding ≤ 10μM |
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
580 |
0.29 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
399 |
0.30 |
Binding ≤ 10μM
|
|
RON-1-E |
Macrophage-stimulating Protein Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM |
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
448 |
0.30 |
Binding ≤ 10μM |
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
294 |
0.30 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK_HUMAN |
Q9UM73
|
ALK Tyrosine Kinase Receptor, Human |
1 |
0.42 |
Binding ≤ 1μM |
|
MET_HUMAN |
P08581
|
Hepatocyte Growth Factor Receptor, Human |
18.3 |
0.36 |
Binding ≤ 1μM |
|
INSR_HUMAN |
P06213
|
Insulin Receptor, Human |
102 |
0.33 |
Binding ≤ 1μM |
|
RON_HUMAN |
Q04912
|
Macrophage-stimulating Protein Receptor, Human |
300 |
0.30 |
Binding ≤ 1μM
|
|
RON_MOUSE |
Q62190
|
Macrophage-stimulating Protein Receptor, Mouse |
80 |
0.33 |
Binding ≤ 1μM |
|
NTRK1_HUMAN |
P04629
|
Nerve Growth Factor Receptor Trk-A, Human |
1 |
0.42 |
Binding ≤ 1μM |
|
NTRK2_HUMAN |
Q16620
|
Neurotrophic Tyrosine Kinase Receptor Type 2, Human |
2 |
0.41 |
Binding ≤ 1μM |
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
24 |
0.36 |
Binding ≤ 1μM |
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
1 |
0.42 |
Binding ≤ 1μM |
|
UFO_HUMAN |
P30530
|
Tyrosine-protein Kinase Receptor UFO, Human |
1 |
0.42 |
Binding ≤ 1μM |
|
TIE2_MOUSE |
Q02858
|
Tyrosine-protein Kinase TIE-2, Mouse |
448 |
0.30 |
Binding ≤ 1μM |
|
TIE2_HUMAN |
Q02763
|
Tyrosine-protein Kinase TIE-2, Human |
448 |
0.30 |
Binding ≤ 1μM
|
|
ALK_HUMAN |
Q9UM73
|
ALK Tyrosine Kinase Receptor, Human |
1 |
0.42 |
Binding ≤ 10μM |
|
MET_HUMAN |
P08581
|
Hepatocyte Growth Factor Receptor, Human |
18.3 |
0.36 |
Binding ≤ 10μM |
|
INSR_HUMAN |
P06213
|
Insulin Receptor, Human |
102 |
0.33 |
Binding ≤ 10μM |
|
RON_MOUSE |
Q62190
|
Macrophage-stimulating Protein Receptor, Mouse |
80 |
0.33 |
Binding ≤ 10μM |
|
RON_HUMAN |
Q04912
|
Macrophage-stimulating Protein Receptor, Human |
300 |
0.30 |
Binding ≤ 10μM
|
|
NTRK1_HUMAN |
P04629
|
Nerve Growth Factor Receptor Trk-A, Human |
1 |
0.42 |
Binding ≤ 10μM |
|
NTRK2_HUMAN |
Q16620
|
Neurotrophic Tyrosine Kinase Receptor Type 2, Human |
2 |
0.41 |
Binding ≤ 10μM |
|
ABL1_MOUSE |
P00520
|
Tyrosine-protein Kinase ABL, Mouse |
1159 |
0.28 |
Binding ≤ 10μM |
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
1159 |
0.28 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
1 |
0.42 |
Binding ≤ 10μM |
|
UFO_HUMAN |
P30530
|
Tyrosine-protein Kinase Receptor UFO, Human |
1 |
0.42 |
Binding ≤ 10μM |
|
TIE2_MOUSE |
Q02858
|
Tyrosine-protein Kinase TIE-2, Mouse |
448 |
0.30 |
Binding ≤ 10μM |
|
TIE2_HUMAN |
Q02763
|
Tyrosine-protein Kinase TIE-2, Human |
448 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
9.09 |
-96.36 |
5 |
6 |
2 |
84 |
452.361 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
8.61 |
-52.37 |
4 |
6 |
1 |
83 |
451.353 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
1.07 |
-45.22 |
3 |
3 |
1 |
34 |
192.286 |
1 |
↓
|
|
|
|
Analogs
-
8699424
-
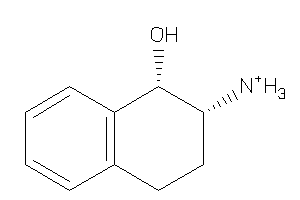
-
8699421
-
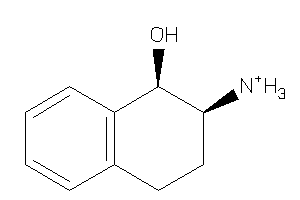
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNMT-1-E |
Phenylethanolamine N-methyltransferase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4600 |
0.62 |
Binding ≤ 10μM
|
|
PNMT-1-E |
Phenylethanolamine N-methyltransferase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.59 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PNMT_BOVIN |
P10938
|
Phenylethanolamine N-methyltransferase, Bovin |
9400 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.50 |
1.12 |
-43.6 |
4 |
2 |
1 |
48 |
164.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.79 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
36 |
0.87 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.84 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.85 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.85 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.85 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.79 |
Binding ≤ 10μM
|
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5400 |
0.61 |
Binding ≤ 10μM
|
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5400 |
0.61 |
Binding ≤ 10μM
|
|
NISCH-2-E |
Nischarin (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5400 |
0.61 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
170 |
0.79 |
Binding ≤ 1μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
36 |
0.87 |
Binding ≤ 1μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
23 |
0.89 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
50 |
0.85 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
50 |
0.85 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
50 |
0.85 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
50 |
0.85 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
180 |
0.79 |
Binding ≤ 1μM
|
|
AOFA_RAT |
P21396
|
Monoamine Oxidase A, Rat |
5400 |
0.61 |
Binding ≤ 10μM
|
|
AOFB_RAT |
P19643
|
Monoamine Oxidase B, Rat |
5400 |
0.61 |
Binding ≤ 10μM
|
|
NISCH_HUMAN |
Q9Y2I1
|
Nischarin, Human |
5400 |
0.61 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
170 |
0.79 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
36 |
0.87 |
Binding ≤ 10μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
23 |
0.89 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
2895 |
0.65 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
2895 |
0.65 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
2895 |
0.65 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
50 |
0.85 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
180 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
3.36 |
-45.89 |
4 |
2 |
1 |
43 |
161.228 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
3.34 |
-179.08 |
0 |
6 |
-3 |
120 |
207.117 |
3 |
↓
|
|
|
|
Analogs
-
44808929
-
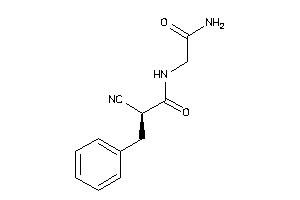
-
44808930
-
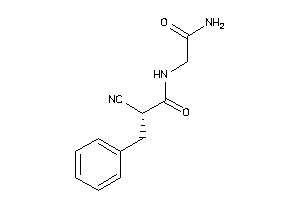
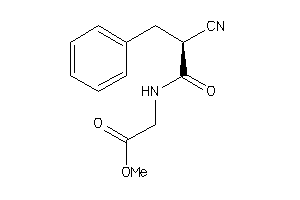
-
44810630
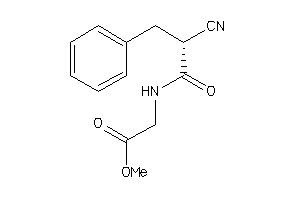
-
-
44810632
-
-
44813367
-
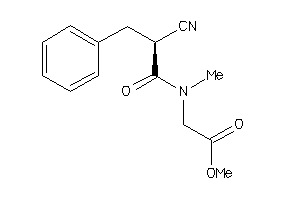
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.53 |
Binding ≤ 10μM
|
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.50 |
Binding ≤ 10μM
|
|
ACE2-1-E |
Angiotensin-converting Enzyme 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.53 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.68 |
Binding ≤ 10μM
|
|
NEP-1-E |
Neprilysin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Functional ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
1800 |
0.47 |
Binding ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
9500 |
0.41 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE_HUMAN |
P12821
|
Angiotensin-converting Enzyme, Human |
140 |
0.56 |
Binding ≤ 1μM
|
|
ACE2_HUMAN |
Q9BYF1
|
Angiotensin-converting Enzyme 2, Human |
420 |
0.53 |
Binding ≤ 1μM
|
|
ECE1_HUMAN |
P42892
|
Endothelin-converting Enzyme 1, Human |
8 |
0.67 |
Binding ≤ 1μM
|
|
NEP_PIG |
P19621
|
Neprilysin, Pig |
6.2 |
0.68 |
Binding ≤ 1μM
|
|
NEP_HUMAN |
P08473
|
Neprilysin, Human |
20 |
0.63 |
Binding ≤ 1μM
|
|
NEP_RAT |
P07861
|
Neprilysin, Rat |
1.5 |
0.73 |
Binding ≤ 1μM
|
|
ACE_HUMAN |
P12821
|
Angiotensin-converting Enzyme, Human |
140 |
0.56 |
Binding ≤ 10μM
|
|
ACE2_HUMAN |
Q9BYF1
|
Angiotensin-converting Enzyme 2, Human |
420 |
0.53 |
Binding ≤ 10μM
|
|
ECE1_HUMAN |
P42892
|
Endothelin-converting Enzyme 1, Human |
8 |
0.67 |
Binding ≤ 10μM
|
|
NEP_RAT |
P07861
|
Neprilysin, Rat |
1.5 |
0.73 |
Binding ≤ 10μM
|
|
NEP_PIG |
P19621
|
Neprilysin, Pig |
6.2 |
0.68 |
Binding ≤ 10μM
|
|
NEP_HUMAN |
P08473
|
Neprilysin, Human |
20 |
0.63 |
Binding ≤ 10μM
|
|
THER_BACTH |
P00800
|
Thermolysin, Bacth |
1800 |
0.47 |
Binding ≤ 10μM
|
|
NEP_RAT |
P07861
|
Neprilysin, Rat |
1.9 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.25 |
5.62 |
-41.1 |
1 |
4 |
-1 |
69 |
252.315 |
6 |
↓
|
|
|
|
Analogs
-
4025720
-
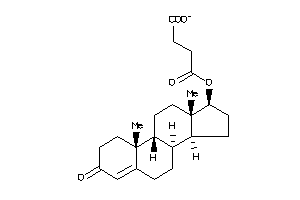
-
4025721
-
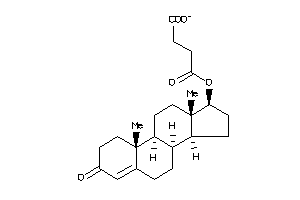
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
2.73 |
-49.55 |
0 |
5 |
-1 |
83 |
387.496 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
3.24 |
-56.49 |
4 |
5 |
1 |
91 |
333.861 |
9 |
↓
|
|
|
|
Analogs
-
41664953
-
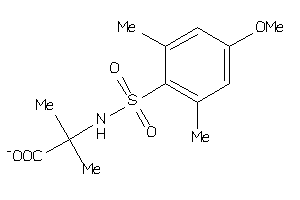
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
1.38 |
-47.5 |
1 |
6 |
-1 |
96 |
258.275 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAKB1-1-E |
AMP-activated Protein Kinase, Beta-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.34 |
Binding ≤ 10μM
|
|
AAKG1-1-E |
AMP-activated Protein Kinase, Gamma-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.34 |
Binding ≤ 10μM
|
|
AAPK2-1-E |
AMP-activated Protein Kinase, Alpha-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
41 |
0.34 |
Binding ≤ 10μM
|
|
BMP4-1-E |
Bone Morphogenetic Protein 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.30 |
Binding ≤ 10μM
|
|
EPHA2-1-E |
Ephrin Type-A Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.37 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.31 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.37 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.37 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAPK2_HUMAN |
P54646
|
AMP-activated Protein Kinase, Alpha-2 Subunit, Human |
41 |
0.34 |
Binding ≤ 1μM
|
|
AAKB1_HUMAN |
Q9Y478
|
AMP-activated Protein Kinase, Beta-1 Subunit, Human |
41 |
0.34 |
Binding ≤ 1μM
|
|
AAKG1_HUMAN |
P54619
|
AMP-activated Protein Kinase, Gamma-1 Subunit, Human |
41 |
0.34 |
Binding ≤ 1μM
|
|
BMP4_HUMAN |
P12644
|
Bone Morphogenetic Protein 4, Human |
430 |
0.30 |
Binding ≤ 1μM
|
|
EPHA2_HUMAN |
P29317
|
Ephrin Type-A Receptor 2, Human |
11 |
0.37 |
Binding ≤ 1μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
11 |
0.37 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
210 |
0.31 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
16 |
0.36 |
Binding ≤ 1μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
1 |
0.42 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
2 |
0.41 |
Binding ≤ 1μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
11 |
0.37 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
4 |
0.39 |
Binding ≤ 1μM
|
|
AAPK2_HUMAN |
P54646
|
AMP-activated Protein Kinase, Alpha-2 Subunit, Human |
41 |
0.34 |
Binding ≤ 10μM
|
|
AAKB1_HUMAN |
Q9Y478
|
AMP-activated Protein Kinase, Beta-1 Subunit, Human |
41 |
0.34 |
Binding ≤ 10μM
|
|
AAKG1_HUMAN |
P54619
|
AMP-activated Protein Kinase, Gamma-1 Subunit, Human |
41 |
0.34 |
Binding ≤ 10μM
|
|
BMP4_HUMAN |
P12644
|
Bone Morphogenetic Protein 4, Human |
430 |
0.30 |
Binding ≤ 10μM
|
|
EPHA2_HUMAN |
P29317
|
Ephrin Type-A Receptor 2, Human |
11 |
0.37 |
Binding ≤ 10μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
11 |
0.37 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
210 |
0.31 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
16 |
0.36 |
Binding ≤ 10μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
1 |
0.42 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
2 |
0.41 |
Binding ≤ 10μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
11 |
0.37 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
12.57 |
-44.33 |
1 |
6 |
1 |
57 |
400.506 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.94 |
10.36 |
-10.49 |
0 |
6 |
0 |
56 |
399.498 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.94 |
13.05 |
-77.93 |
2 |
6 |
2 |
58 |
401.514 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-1-E |
Dipeptidyl Peptidase IV (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.41 |
Binding ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
-1.38 |
-69.61 |
3 |
6 |
1 |
89 |
360.441 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102317-1-O |
Brain (cluster #1 Of 2), Other |
Other |
4770 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
2.4 |
-49.65 |
1 |
5 |
-1 |
79 |
223.204 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
5.18 |
-48.91 |
1 |
5 |
-1 |
82 |
272.078 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.98 |
3.21 |
-12.33 |
2 |
5 |
0 |
79 |
273.086 |
4 |
↓
|
|
|
|
Analogs
-
12832834
-
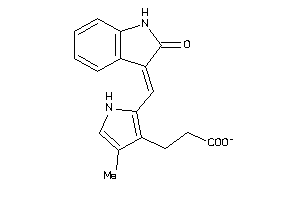
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.40 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.49 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.35 |
Functional ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
50 |
0.46 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
30 |
0.48 |
Binding ≤ 1μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
510 |
0.40 |
Binding ≤ 1μM
|
|
VGFR1_MOUSE |
P35969
|
Vascular Endothelial Growth Factor Receptor 1, Mouse |
400 |
0.41 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
20 |
0.49 |
Binding ≤ 1μM
|
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
10000 |
0.32 |
Binding ≤ 10μM
|
|
FGFR1_MOUSE |
P16092
|
Fibroblast Growth Factor Receptor 1, Mouse |
10000 |
0.32 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
510 |
0.40 |
Binding ≤ 10μM
|
|
VGFR1_MOUSE |
P35969
|
Vascular Endothelial Growth Factor Receptor 1, Mouse |
400 |
0.41 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
20 |
0.49 |
Binding ≤ 10μM
|
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
2800 |
0.35 |
Functional ≤ 10μM
|
|
Z81057 |
Z81057
|
HUVEC (Umbilical Vein Endothelial Cells) |
2800 |
0.35 |
Functional ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
50 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-0.6 |
-51.65 |
2 |
5 |
-1 |
88 |
295.318 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-4.73 |
-35.44 |
5 |
8 |
1 |
122 |
272.281 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.19 |
-3.91 |
-60.08 |
4 |
6 |
-1 |
115 |
209.181 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.66 |
2.77 |
-49.17 |
2 |
8 |
-1 |
135 |
239.163 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
46 |
0.38 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.29 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.27 |
Binding ≤ 10μM
|
|
CSK22-1-E |
Casein Kinase II Alpha (prime) (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.27 |
Binding ≤ 10μM
|
|
CSK2B-1-E |
Casein Kinase II Beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5400 |
0.27 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.40 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.40 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.40 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.27 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
360 |
0.33 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
580 |
0.32 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.34 |
Binding ≤ 10μM
|
|
ROCK1-1-E |
Rho-associated Protein Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.37 |
Binding ≤ 10μM
|
|
Z80800-1-O |
MIA PaCa-2 (Pancreatic Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.33 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 1μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 1μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 1μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
46 |
0.38 |
Binding ≤ 1μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
110 |
0.36 |
Binding ≤ 1μM
|
|
MK01_HUMAN |
P28482
|
MAP Kinase ERK2, Human |
260 |
0.34 |
Binding ≤ 1μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
360 |
0.33 |
Binding ≤ 1μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
79 |
0.37 |
Binding ≤ 1μM
|
|
KS6A3_HUMAN |
P51812
|
Ribosomal Protein S6 Kinase Alpha 3, Human |
580 |
0.32 |
Binding ≤ 1μM
|
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
11 |
0.41 |
Binding ≤ 1μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
16 |
0.40 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
5400 |
0.27 |
Binding ≤ 10μM
|
|
CSK22_HUMAN |
P19784
|
Casein Kinase II Alpha (prime), Human |
5400 |
0.27 |
Binding ≤ 10μM
|
|
CSK2B_HUMAN |
P67870
|
Casein Kinase II Beta, Human |
5400 |
0.27 |
Binding ≤ 10μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
46 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
110 |
0.36 |
Binding ≤ 10μM
|
|
MK01_HUMAN |
P28482
|
MAP Kinase ERK2, Human |
260 |
0.34 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
1100 |
0.31 |
Binding ≤ 10μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
360 |
0.33 |
Binding ≤ 10μM
|
|
KPCG_HUMAN |
P05129
|
Protein Kinase C Gamma, Human |
1200 |
0.31 |
Binding ≤ 10μM
|
|
ROCK1_HUMAN |
Q13464
|
Rho-associated Protein Kinase 1, Human |
79 |
0.37 |
Binding ≤ 10μM
|
|
KS6A3_HUMAN |
P51812
|
Ribosomal Protein S6 Kinase Alpha 3, Human |
580 |
0.32 |
Binding ≤ 10μM
|
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
11 |
0.41 |
Binding ≤ 10μM
|
|
CHK1_HUMAN |
O14757
|
Serine/threonine-protein Kinase Chk1, Human |
2600 |
0.29 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
5800 |
0.27 |
Binding ≤ 10μM
|
|
Z80800 |
Z80800
|
MIA PaCa-2 (Pancreatic Carcinoma Cells) |
400 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
7.58 |
-52.57 |
4 |
5 |
1 |
78 |
359.453 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.21 |
5.96 |
-91.78 |
7 |
8 |
2 |
122 |
470.524 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.42 |
7.29 |
-9.63 |
3 |
5 |
0 |
77 |
358.445 |
6 |
↓
|
|
|
|
Analogs
-
4677
-
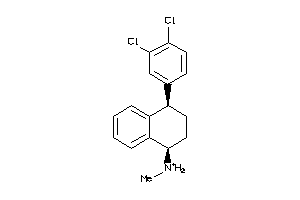
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.47 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
825 |
0.43 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.46 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.46 |
Functional ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.45 |
Functional ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.51 |
Functional ≤ 10μM
|
|
CP2CJ-1-E |
Cytochrome P450 2C19 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.46 |
ADME/T ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
800 |
0.43 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.38 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
60 |
0.51 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
25 |
0.53 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
260 |
0.46 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
159 |
0.48 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
420 |
0.45 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
0.85 |
0.63 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.29 |
0.67 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
260 |
0.46 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
25 |
0.53 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
420 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
159 |
0.48 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
0.85 |
0.63 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.29 |
0.67 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
520 |
0.44 |
Functional ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
720 |
0.43 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.35 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1200 |
0.41 |
Functional ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
70 |
0.50 |
Functional ≤ 10μM
|
|
CP2CJ_HUMAN |
P33261
|
Cytochrome P450 2C19, Human |
310 |
0.46 |
ADME/T ≤ 10μM
|
|
CP2D6_HUMAN |
P10635
|
Cytochrome P450 2D6, Human |
1400 |
0.41 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
800 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
10.12 |
-46.17 |
2 |
1 |
1 |
17 |
307.244 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
512 |
0.31 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1750 |
0.29 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
1750 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
9.41 |
-46.52 |
0 |
5 |
-1 |
66 |
416.454 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.31 |
9.19 |
-11.34 |
1 |
5 |
0 |
64 |
417.462 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
398 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
8.57 |
-33.96 |
1 |
2 |
1 |
23 |
351.651 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.26 |
7.66 |
-6.24 |
0 |
2 |
0 |
22 |
350.643 |
2 |
↓
|
|
|
|
Analogs
-
4027073
-
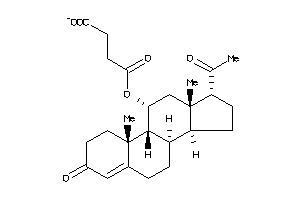
-
4027074
-
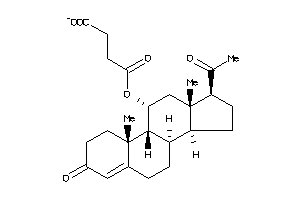
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
11.79 |
-51.99 |
0 |
6 |
-1 |
101 |
429.533 |
6 |
↓
|
|
|
|
Analogs
-
1530638
-
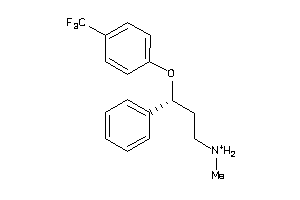
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
33 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
130 |
0.44 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
605 |
0.40 |
Binding ≤ 1μM
|
|
SC6A3_MOUSE |
Q61327
|
Dopamine Transporter, Mouse |
10.8 |
0.51 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
784 |
0.39 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
457.08819 |
0.40 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
143 |
0.44 |
Binding ≤ 1μM
|
|
SC6A2_MOUSE |
O55192
|
Norepinephrine Transporter, Mouse |
85 |
0.45 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
240 |
0.42 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
2 |
0.55 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.72 |
0.58 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
473 |
0.40 |
Binding ≤ 1μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
130 |
0.44 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
2620 |
0.36 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
784 |
0.39 |
Binding ≤ 10μM
|
|
SC6A3_MOUSE |
Q61327
|
Dopamine Transporter, Mouse |
10.8 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
1900 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1513.56125 |
0.37 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
7300 |
0.33 |
Binding ≤ 10μM
|
|
SC6A2_MOUSE |
O55192
|
Norepinephrine Transporter, Mouse |
85 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
143 |
0.44 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
1020 |
0.38 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
2 |
0.55 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.72 |
0.58 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
473 |
0.40 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
33 |
0.48 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
193 |
0.43 |
Functional ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
130 |
0.44 |
Functional ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
7000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-43.81 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.33 |
-61.83 |
1 |
5 |
-1 |
78 |
340.399 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.34 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.35 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.33 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.33 |
Functional ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
103 |
0.32 |
Functional ≤ 10μM
|
|
PPARD-1-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.33 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
-0.53 |
-55.12 |
0 |
6 |
-1 |
84 |
436.509 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
5.79 |
-15.08 |
1 |
6 |
0 |
93 |
362.432 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
3.78 |
5.7 |
-14.92 |
1 |
6 |
0 |
93 |
362.432 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
3.79 |
-60.86 |
0 |
6 |
-1 |
96 |
361.424 |
3 |
↓
|
|
|
|
Analogs
-
89503
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.36 |
3.95 |
-39.31 |
7 |
7 |
1 |
118 |
238.271 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.36 |
3.98 |
-7.96 |
6 |
7 |
0 |
116 |
237.263 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.24 |
3.7 |
-101.4 |
8 |
7 |
2 |
122 |
239.279 |
5 |
↓
|
|
|
|
Analogs
-
73041
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
6.31 |
-29.27 |
7 |
5 |
1 |
100 |
280.661 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.57 |
4.66 |
-9.87 |
6 |
5 |
0 |
103 |
279.653 |
3 |
↓
|
|
|
|
Analogs
-
1530637
-
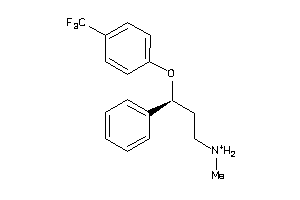
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
31 |
0.48 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
21 |
0.49 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
130 |
0.44 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
605 |
0.40 |
Binding ≤ 1μM
|
|
SC6A3_MOUSE |
Q61327
|
Dopamine Transporter, Mouse |
10.8 |
0.51 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
784 |
0.39 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
457.08819 |
0.40 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
143 |
0.44 |
Binding ≤ 1μM
|
|
SC6A2_MOUSE |
O55192
|
Norepinephrine Transporter, Mouse |
85 |
0.45 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
240 |
0.42 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
15 |
0.50 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
2 |
0.55 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.72 |
0.58 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
473 |
0.40 |
Binding ≤ 1μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
130 |
0.44 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
2620 |
0.36 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
1920 |
0.36 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
784 |
0.39 |
Binding ≤ 10μM
|
|
SC6A3_MOUSE |
Q61327
|
Dopamine Transporter, Mouse |
10.8 |
0.51 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
1900 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1513.56125 |
0.37 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
7300 |
0.33 |
Binding ≤ 10μM
|
|
SC6A2_MOUSE |
O55192
|
Norepinephrine Transporter, Mouse |
85 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
143 |
0.44 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
1020 |
0.38 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
15 |
0.50 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
2 |
0.55 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
0.72 |
0.58 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
473 |
0.40 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
21 |
0.49 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
193 |
0.43 |
Functional ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
130 |
0.44 |
Functional ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
7000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-44.31 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
175214
-
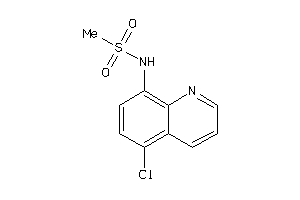
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
1.75 |
-47.86 |
0 |
4 |
-1 |
61 |
221.261 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.13 |
2.24 |
-36.25 |
1 |
4 |
0 |
62 |
222.269 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.13 |
1.71 |
-11.9 |
1 |
4 |
0 |
59 |
222.269 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
5.33 |
-52.38 |
0 |
6 |
-1 |
96 |
369.422 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.31 |
7.06 |
-16.88 |
1 |
6 |
0 |
89 |
370.43 |
5 |
↓
|
|
|
|
Analogs
-
17044325
-
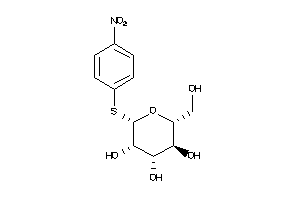
-
17044328
-
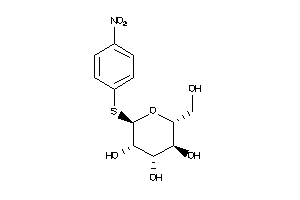
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.01 |
-3.71 |
-9.33 |
4 |
8 |
0 |
136 |
317.319 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.43 |
2.08 |
-118.87 |
1 |
6 |
-2 |
118 |
224.168 |
4 |
↓
|
|
|
|
Analogs
-
6357278
-
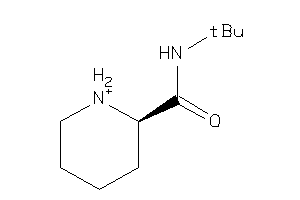
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.28 |
-2.02 |
-35 |
3 |
3 |
1 |
45 |
185.291 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
4.88 |
-65.92 |
3 |
8 |
-1 |
136 |
440.472 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
-2.74 |
-51.68 |
1 |
7 |
-1 |
104 |
403.201 |
5 |
↓
|
|
|
|
Analogs
-
1731932
-
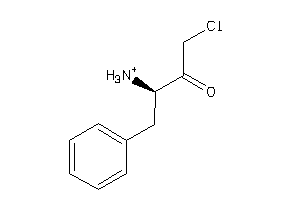
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
4.54 |
-46.3 |
3 |
2 |
1 |
45 |
198.673 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.23 |
-1.23 |
-42.81 |
3 |
4 |
0 |
84 |
269.3 |
5 |
↓
|
|
|
|
Analogs
-
6251368
-
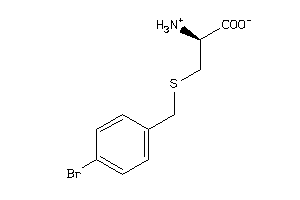
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
-3.08 |
-37.49 |
3 |
3 |
0 |
67 |
290.182 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
0.36 |
-42.84 |
2 |
2 |
1 |
21 |
239.342 |
3 |
↓
|
|
|
|
Analogs
-
38208359
-
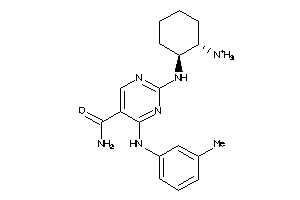
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
-5.74 |
-53.58 |
7 |
7 |
1 |
120 |
341.439 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-2-E |
Poly [ADP-ribose] Polymerase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.49 |
Binding ≤ 10μM
|
|
PARP3-1-E |
Poly [ADP-ribose] Polymerase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
-0.77 |
-51.13 |
3 |
5 |
1 |
66 |
296.35 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
-0.75 |
-47.57 |
2 |
4 |
-1 |
81 |
167.14 |
2 |
↓
|
|