|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
35 |
0.33 |
Binding ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80119-1-O |
Detroit 98 (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
5 |
0.36 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7900 |
0.22 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2050 |
0.25 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z81131-1-O |
L Cells (Fibroblasts) (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
4.48 |
-122.21 |
6 |
13 |
-2 |
225 |
438.404 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.21 |
4.93 |
-127.62 |
7 |
13 |
-1 |
226 |
439.412 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
33 |
0.34 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1680 |
0.26 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1680 |
0.26 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1680 |
0.26 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
210 |
0.30 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Binding ≤ 10μM
|
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Functional ≤ 10μM
|
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
3 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.40 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
3.4 |
0.38 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.37 |
0.43 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 1μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.33 |
0.43 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.66 |
0.41 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
210 |
0.30 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
12 |
0.36 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
0.45 |
0.42 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
0.45 |
0.42 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
2.6 |
0.39 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
11 |
0.36 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10 |
0.36 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
0.51 |
0.42 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
33 |
0.34 |
Binding ≤ 1μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
56 |
0.33 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
0.6 |
0.42 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
0.2 |
0.44 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
0.39 |
0.42 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
0.2 |
0.44 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
0.39 |
0.42 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
5 |
0.37 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.40 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.37 |
0.43 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
3.4 |
0.38 |
Binding ≤ 10μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.33 |
0.43 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
1.8 |
0.39 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.66 |
0.41 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
1680 |
0.26 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
1680 |
0.26 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
1680 |
0.26 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
210 |
0.30 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
12 |
0.36 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
0.45 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
0.45 |
0.42 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
2.6 |
0.39 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
11 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10 |
0.36 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
0.51 |
0.42 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
33 |
0.34 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
56 |
0.33 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
0.6 |
0.42 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
0.2 |
0.44 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
0.39 |
0.42 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
0.2 |
0.44 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
0.39 |
0.42 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
5 |
0.37 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
14 |
0.35 |
Functional ≤ 10μM
|
|
KCNA5_HUMAN |
P22460
|
Voltage-gated Potassium Channel Subunit Kv1.5, Human |
2000 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
13.19 |
-49.8 |
2 |
5 |
1 |
42 |
441.958 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.2 |
-69.16 |
2 |
3 |
0 |
57 |
337.463 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.70 |
11.23 |
-43.27 |
3 |
3 |
1 |
54 |
338.471 |
8 |
↓
|
|
|
|
Analogs
-
7996519
-
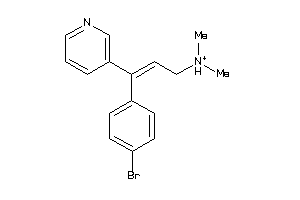
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
10.2 |
-42.99 |
1 |
2 |
1 |
17 |
318.238 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
7.54 |
-4.17 |
0 |
2 |
0 |
16 |
317.23 |
4 |
↓
|
|
|
|
Analogs
-
8099545
-
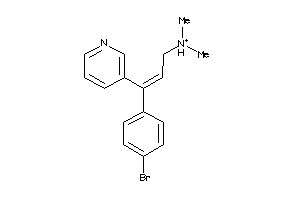
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
10.2 |
-41.9 |
1 |
2 |
1 |
17 |
318.238 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.37 |
7.55 |
-4.86 |
0 |
2 |
0 |
16 |
317.23 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4300 |
0.38 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
4300 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
8.51 |
-46.08 |
0 |
4 |
-1 |
66 |
330.168 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.20 |
2.84 |
-25.7 |
7 |
5 |
1 |
104 |
158.229 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-0.20 |
2.71 |
-4.62 |
7 |
5 |
0 |
104 |
158.229 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.06 |
4.04 |
-5.71 |
6 |
5 |
0 |
98 |
157.221 |
6 |
↓
|
|
|
|
Analogs
-
32264981
-
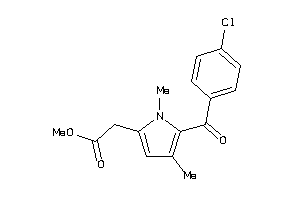
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
1630 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
9.6 |
-44.32 |
0 |
4 |
-1 |
62 |
290.726 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
10.55 |
-39.34 |
1 |
2 |
1 |
8 |
287.452 |
4 |
↓
|
|
|
|
Analogs
-
1532185
-
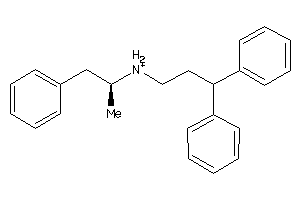
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
209 |
0.37 |
Binding ≤ 10μM |
|
CAC1D-2-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
209 |
0.37 |
Binding ≤ 10μM |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
209 |
0.37 |
Binding ≤ 1μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
209 |
0.37 |
Binding ≤ 1μM
|
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
209 |
0.37 |
Binding ≤ 10μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
209 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
14.22 |
-40.29 |
2 |
1 |
1 |
17 |
330.495 |
8 |
↓
|
|
|
|
Analogs
-
1532186
-
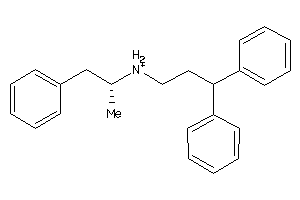
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
209 |
0.37 |
Binding ≤ 10μM |
|
CAC1D-2-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
209 |
0.37 |
Binding ≤ 10μM |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 1μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
209 |
0.37 |
Binding ≤ 1μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
209 |
0.37 |
Binding ≤ 1μM
|
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
300 |
0.37 |
Binding ≤ 10μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
209 |
0.37 |
Binding ≤ 10μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
209 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
14.22 |
-39.91 |
2 |
1 |
1 |
17 |
330.495 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
11.68 |
-35.93 |
1 |
4 |
1 |
40 |
336.496 |
13 |
↓
|
|
|
|
Analogs
-
1733848
-
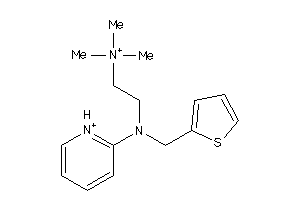
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
10.02 |
-90.68 |
2 |
3 |
2 |
22 |
263.41 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.47 |
7.16 |
-4.25 |
0 |
3 |
0 |
19 |
261.394 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.47 |
9.64 |
-41.53 |
1 |
3 |
1 |
21 |
262.402 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.50 |
2.74 |
-72.05 |
0 |
7 |
-1 |
87 |
310.355 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.50 |
3.49 |
-38.8 |
1 |
7 |
0 |
89 |
311.363 |
4 |
↓
|
|
|
|
Analogs
-
70
-
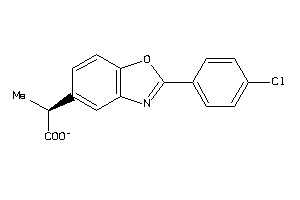
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
-0.61 |
-46.47 |
0 |
4 |
-1 |
66 |
300.721 |
3 |
↓
|
|
|
|
Analogs
-
1872015
-
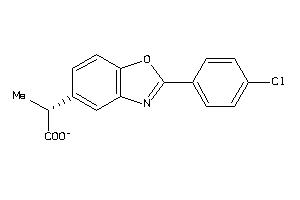
-
33420303
-
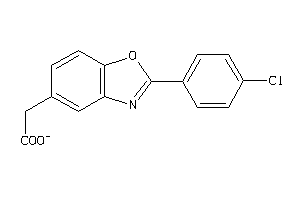
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
-0.61 |
-46.43 |
0 |
4 |
-1 |
66 |
300.721 |
3 |
↓
|
|
|
|
Analogs
-
34007650
-
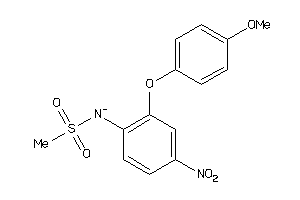
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3760 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-8-E |
Cyclooxygenase-2 (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
400 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
4.93 |
-37.19 |
0 |
7 |
-1 |
103 |
307.307 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.81 |
4.82 |
-11.73 |
1 |
7 |
0 |
101 |
308.315 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1A1_PIG |
P50578
|
Aldehyde Reductase, Pig |
540 |
0.37 |
Binding ≤ 1μM
|
|
AK1A1_HUMAN |
P14550
|
Aldehyde Reductase, Human |
720 |
0.36 |
Binding ≤ 1μM
|
|
AK1BA_HUMAN |
O60218
|
Aldo-keto Reductase Family 1 Member B10, Human |
11.6 |
0.46 |
Binding ≤ 1μM
|
|
ALDR_PIG |
P80276
|
Aldose Reductase, Pig |
15 |
0.46 |
Binding ≤ 1μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
33 |
0.44 |
Binding ≤ 1μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
10 |
0.47 |
Binding ≤ 1μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1000 |
0.35 |
Binding ≤ 1μM
|
|
PPBT_RAT |
P08289
|
Alkaline Phosphatase Tissue-nonspecific, Rat |
27.2 |
0.44 |
Binding ≤ 1μM
|
|
AK1A1_PIG |
P50578
|
Aldehyde Reductase, Pig |
2190 |
0.33 |
Binding ≤ 10μM
|
|
AK1A1_HUMAN |
P14550
|
Aldehyde Reductase, Human |
1940 |
0.33 |
Binding ≤ 10μM
|
|
AK1A1_RAT |
P51635
|
Aldehyde Reductase, Rat |
1210 |
0.35 |
Binding ≤ 10μM
|
|
AK1BA_HUMAN |
O60218
|
Aldo-keto Reductase Family 1 Member B10, Human |
11.6 |
0.46 |
Binding ≤ 10μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
33 |
0.44 |
Binding ≤ 10μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
10 |
0.47 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1000 |
0.35 |
Binding ≤ 10μM
|
|
ALDR_PIG |
P80276
|
Aldose Reductase, Pig |
15 |
0.46 |
Binding ≤ 10μM
|
|
PPBT_RAT |
P08289
|
Alkaline Phosphatase Tissue-nonspecific, Rat |
27.2 |
0.44 |
Binding ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
15 |
0.46 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
10.02 |
-52.28 |
0 |
4 |
-1 |
53 |
356.345 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
5065159
-
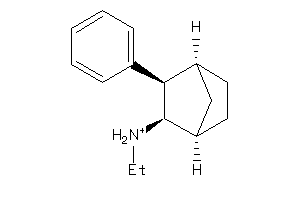
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
0.2 |
-37.63 |
2 |
1 |
1 |
16 |
216.348 |
3 |
↓
|
|
|
|
Analogs
-
5065160
-
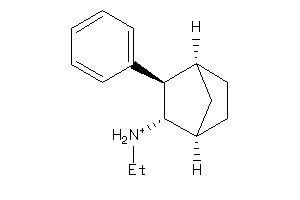
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
1.23 |
-30.24 |
2 |
1 |
1 |
16 |
216.348 |
3 |
↓
|
|
|
|
Analogs
-
9133461
-
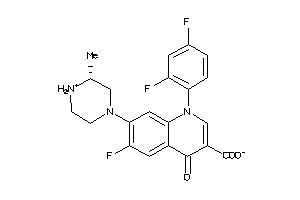
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
11.86 |
-94.26 |
2 |
6 |
0 |
82 |
417.387 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.37 |
10.62 |
-61.88 |
1 |
6 |
-1 |
77 |
416.379 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.38 |
9.85 |
-78.71 |
3 |
6 |
1 |
85 |
418.395 |
2 |
↓
|
|
|
|
Analogs
-
2004103
-
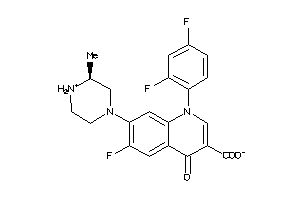
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
0.76 |
-96.96 |
2 |
6 |
0 |
82 |
417.387 |
3 |
↓
|
|
|
|
Analogs
-
607970
-
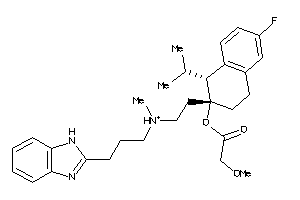
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1340 |
0.23 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
860 |
0.24 |
Binding ≤ 10μM |
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2-4-E |
HERG (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
806 |
0.24 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
416 |
0.25 |
Functional ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.24 |
Functional ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.29 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.23 |
Functional ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.22 |
Functional ≤ 10μM
|
|
MDR3-2-E |
P-glycoprotein 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.20 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
130 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
1 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
398 |
0.25 |
Functional ≤ 10μM
|
|
Z80107-2-O |
COS-7 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
1430 |
0.23 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
190 |
0.26 |
Binding ≤ 1μM
|
|
CAC1G_HUMAN |
O43497
|
Voltage-gated T-type Calcium Channel Alpha-1G Subunit, Human |
1000 |
0.23 |
Binding ≤ 1μM
|
|
CAC1H_HUMAN |
O95180
|
Voltage-gated T-type Calcium Channel Alpha-1H Subunit, Human |
1000 |
0.23 |
Binding ≤ 1μM
|
|
CAC1I_HUMAN |
Q9P0X4
|
Voltage-gated T-type Calcium Channel Alpha-1I Subunit, Human |
1000 |
0.23 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1340 |
0.23 |
Binding ≤ 10μM |
|
CAC1B_HUMAN |
Q00975
|
Voltage-gated N-type Calcium Channel Alpha-1B Subunit, Human |
1340 |
0.23 |
Binding ≤ 10μM
|
|
CAC1G_HUMAN |
O43497
|
Voltage-gated T-type Calcium Channel Alpha-1G Subunit, Human |
1000 |
0.23 |
Binding ≤ 10μM
|
|
CAC1H_HUMAN |
O95180
|
Voltage-gated T-type Calcium Channel Alpha-1H Subunit, Human |
1000 |
0.23 |
Binding ≤ 10μM
|
|
CAC1I_HUMAN |
Q9P0X4
|
Voltage-gated T-type Calcium Channel Alpha-1I Subunit, Human |
1000 |
0.23 |
Binding ≤ 10μM
|
|
Z80107 |
Z80107
|
COS-7 (Kidney Cells) |
1430 |
0.23 |
Functional ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1430 |
0.23 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1500 |
0.23 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
10000 |
0.19 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
7400 |
0.20 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
398.107171 |
0.25 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
0.873 |
0.35 |
Functional ≤ 10μM
|
|
CAC1B_HUMAN |
Q00975
|
Voltage-gated N-type Calcium Channel Alpha-1B Subunit, Human |
416 |
0.25 |
Functional ≤ 10μM
|
|
CAC1G_HUMAN |
O43497
|
Voltage-gated T-type Calcium Channel Alpha-1G Subunit, Human |
1340 |
0.23 |
Functional ≤ 10μM
|
|
CAC1H_HUMAN |
O95180
|
Voltage-gated T-type Calcium Channel Alpha-1H Subunit, Human |
130 |
0.27 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
130 |
0.27 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
14.86 |
-54.6 |
2 |
6 |
1 |
69 |
496.647 |
12 |
↓
|
|
|
|
Analogs
-
597290
-
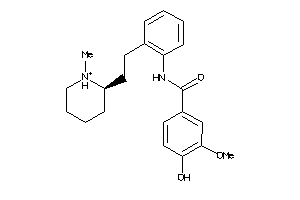
-
607849
-
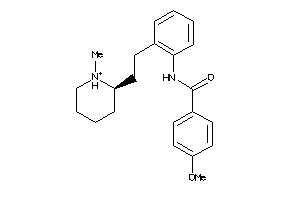
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
11.11 |
-48.47 |
2 |
4 |
1 |
43 |
353.486 |
6 |
↓
|
|
|
|
Analogs
-
4213101
-
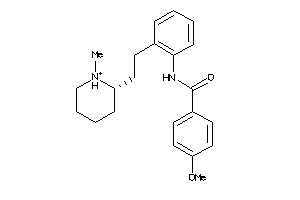
-
597290
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
1.53 |
-49.51 |
2 |
4 |
1 |
42 |
353.486 |
6 |
↓
|
|
|
|
Analogs
-
8830624
-
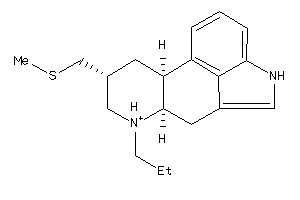
-
33884202
-
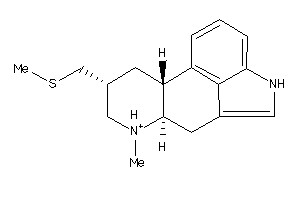
-
1909
-
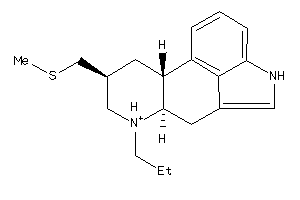
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.34 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
0.1 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
11.27 |
-39.66 |
2 |
2 |
1 |
20 |
315.506 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.97 |
9.08 |
-5.67 |
1 |
2 |
0 |
19 |
314.498 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM |
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.26 |
Binding ≤ 10μM
|
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
360 |
0.27 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.27 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
137 |
0.29 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
360 |
0.27 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
570 |
0.26 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
360 |
0.27 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
5.8 |
0.35 |
Binding ≤ 1μM |
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
137 |
0.29 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
200 |
0.28 |
Binding ≤ 1μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
360 |
0.27 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
2380 |
0.24 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
570 |
0.26 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
360 |
0.27 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
5.8 |
0.35 |
Binding ≤ 10μM |
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
137 |
0.29 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
200 |
0.28 |
Binding ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
8000 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
14.82 |
-48.69 |
1 |
7 |
1 |
57 |
471.025 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.06 |
12.61 |
-18.23 |
0 |
7 |
0 |
56 |
470.017 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
4.2 |
-11.31 |
1 |
4 |
0 |
74 |
329.44 |
0 |
↓
|
|
|
|
Analogs
-
531272
-
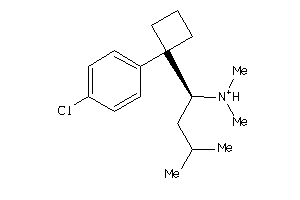
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.53 |
-34.64 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
Analogs
-
4759
-
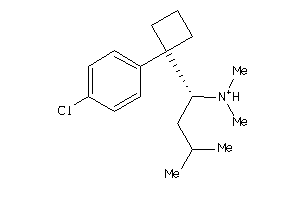
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.74 |
-33.26 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
5.57 |
-58.62 |
6 |
6 |
1 |
95 |
302.402 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.33 |
5.67 |
-20.15 |
5 |
6 |
0 |
93 |
301.394 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.33 |
6.02 |
-105.65 |
7 |
6 |
2 |
96 |
303.41 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
8.94 |
-44.5 |
2 |
5 |
1 |
55 |
295.366 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.25 |
8.94 |
-43.84 |
2 |
5 |
1 |
55 |
295.366 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
8.47 |
-18.57 |
1 |
5 |
0 |
54 |
294.358 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
5.93 |
-7.01 |
7 |
5 |
0 |
100 |
206.273 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.17 |
4.94 |
-28.01 |
7 |
5 |
1 |
104 |
206.273 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
5.35 |
-6.17 |
6 |
5 |
0 |
98 |
205.265 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.26 |
2.45 |
-48.8 |
3 |
6 |
1 |
89 |
462.566 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.26 |
2.37 |
-48.02 |
3 |
6 |
1 |
89 |
462.566 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
-1.72 |
-51.3 |
2 |
7 |
-1 |
109 |
279.301 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.38 |
-1.22 |
-13.36 |
3 |
7 |
0 |
107 |
280.309 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8220 |
0.21 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
SSR5-1-E |
Somatostatin Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4070 |
0.22 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.27 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
734 |
0.25 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
8000 |
0.21 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6500 |
0.21 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.33 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
1 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGTB1_HUMAN |
P53609
|
Geranylgeranyl Transferase Type I Beta Subunit, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
0.9 |
0.37 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
3 |
0.35 |
Binding ≤ 1μM
|
|
FNTB_BOVIN |
P49355
|
Protein Farnesyltransferase Beta Subunit, Bovin |
2 |
0.36 |
Binding ≤ 1μM
|
|
FNTA_BOVIN |
P29702
|
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit, Bovin |
2 |
0.36 |
Binding ≤ 1μM
|
|
PGTB1_HUMAN |
P53609
|
Geranylgeranyl Transferase Type I Beta Subunit, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
0.9 |
0.37 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
3 |
0.35 |
Binding ≤ 10μM
|
|
FNTB_BOVIN |
P49355
|
Protein Farnesyltransferase Beta Subunit, Bovin |
2 |
0.36 |
Binding ≤ 10μM
|
|
FNTA_BOVIN |
P29702
|
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit, Bovin |
2 |
0.36 |
Binding ≤ 10μM
|
|
SSR5_HUMAN |
P35346
|
Somatostatin Receptor 5, Human |
4070 |
0.22 |
Binding ≤ 10μM
|
|
Z80120 |
Z80120
|
DLD-1 (Colon Adenocarcinoma Cells) |
10 |
0.33 |
Functional ≤ 10μM
|
|
Z80936 |
Z80936
|
HEK293 (Embryonic Kidney Fibroblasts) |
0.9 |
0.37 |
Functional ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
0.9 |
0.37 |
Functional ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
3300 |
0.23 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1300 |
0.24 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
1700 |
0.24 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
1300 |
0.24 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
734 |
0.25 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
6500 |
0.21 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
3300 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
17.29 |
-53.86 |
2 |
5 |
1 |
44 |
459.589 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.71 |
17.71 |
-105.8 |
3 |
5 |
2 |
45 |
460.597 |
8 |
↓
|
|
|
|
Analogs
-
4215367
-
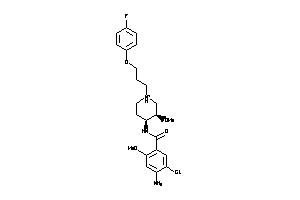
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.36 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
95 |
0.31 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
29 |
0.33 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.23 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
227 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7400 |
0.22 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Functional ≤ 10μM |
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.32 |
Functional ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.36 |
Functional ≤ 10μM
|
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
680 |
0.27 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
110 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
110 |
0.30 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
30 |
0.33 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
30 |
0.33 |
Binding ≤ 1μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
227 |
0.29 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
227 |
0.29 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
227 |
0.29 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10 |
0.35 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
7.2 |
0.36 |
Binding ≤ 1μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 1μM
|
|
5HT3A_HUMAN |
P46098
|
Serotonin 3a (5-HT3a) Receptor, Human |
134 |
0.30 |
Binding ≤ 1μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
134 |
0.30 |
Binding ≤ 1μM
|
|
5HT3A_CAVPO |
O70212
|
Serotonin 3a (5-HT3a) Receptor, Guinea Pig |
134 |
0.30 |
Binding ≤ 1μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
134 |
0.30 |
Binding ≤ 1μM
|
|
5HT3B_HUMAN |
O95264
|
Serotonin 3b (5-HT3b) Receptor, Human |
134 |
0.30 |
Binding ≤ 1μM
|
|
5HT3C_HUMAN |
Q8WXA8
|
Serotonin 3c (5-HT3c) Receptor, Human |
152 |
0.30 |
Binding ≤ 1μM
|
|
5HT3D_HUMAN |
Q70Z44
|
Serotonin 3d (5-HT3d) Receptor, Human |
152 |
0.30 |
Binding ≤ 1μM
|
|
5HT3E_HUMAN |
A5X5Y0
|
Serotonin 3e (5-HT3e) Receptor, Human |
152 |
0.30 |
Binding ≤ 1μM
|
|
5HT4R_HUMAN |
Q13639
|
Serotonin 4 (5-HT4) Receptor, Human |
14.3 |
0.34 |
Binding ≤ 1μM
|
|
5HT4R_CAVPO |
O70528
|
Serotonin 4 (5-HT4) Receptor, Guinea Pig |
17 |
0.34 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
110 |
0.30 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
30 |
0.33 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
4500 |
0.23 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
4500 |
0.23 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
1700 |
0.25 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
1700 |
0.25 |
Binding ≤ 10μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
1700 |
0.25 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
227 |
0.29 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
227 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
4470 |
0.23 |
Binding ≤ 10μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
7.2 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
6.1 |
0.36 |
Binding ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
134 |
0.30 |
Binding ≤ 10μM
|
|
5HT3A_CAVPO |
O70212
|
Serotonin 3a (5-HT3a) Receptor, Guinea Pig |
134 |
0.30 |
Binding ≤ 10μM
|
|
5HT3A_HUMAN |
P46098
|
Serotonin 3a (5-HT3a) Receptor, Human |
134 |
0.30 |
Binding ≤ 10μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
134 |
0.30 |
Binding ≤ 10μM
|
|
5HT3B_HUMAN |
O95264
|
Serotonin 3b (5-HT3b) Receptor, Human |
134 |
0.30 |
Binding ≤ 10μM
|
|
5HT3C_HUMAN |
Q8WXA8
|
Serotonin 3c (5-HT3c) Receptor, Human |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT3D_HUMAN |
Q70Z44
|
Serotonin 3d (5-HT3d) Receptor, Human |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT3E_HUMAN |
A5X5Y0
|
Serotonin 3e (5-HT3e) Receptor, Human |
152 |
0.30 |
Binding ≤ 10μM
|
|
5HT4R_CAVPO |
O70528
|
Serotonin 4 (5-HT4) Receptor, Guinea Pig |
17 |
0.34 |
Binding ≤ 10μM
|
|
5HT4R_HUMAN |
Q13639
|
Serotonin 4 (5-HT4) Receptor, Human |
14.3 |
0.34 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
6.5 |
0.36 |
Functional ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
1500 |
0.25 |
Functional ≤ 10μM
|
|
5HT4R_RAT |
Q62758
|
Serotonin 4 (5-HT4) Receptor, Rat |
54.7 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.67 |
-54.72 |
4 |
7 |
1 |
87 |
466.961 |
9 |
↓
|
|
|
|
Analogs
-
294
-
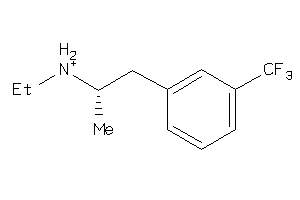
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
7.69 |
-38.85 |
2 |
1 |
1 |
17 |
232.269 |
5 |
↓
|
|
|
|
Analogs
-
1612
-
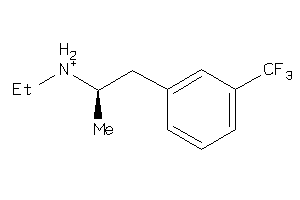
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
7.61 |
-39.24 |
2 |
1 |
1 |
17 |
232.269 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
5851655
-
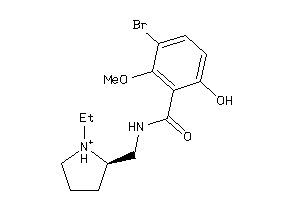
-
5851780
-
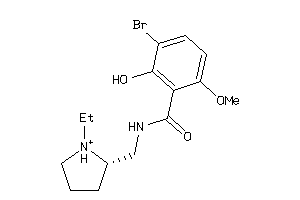
-
5851784
-
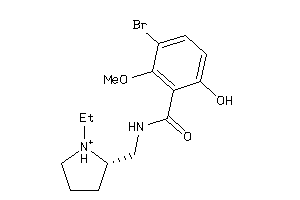
-
597536
-
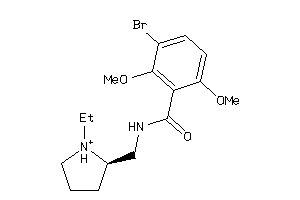
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1570 |
0.37 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4603 |
0.34 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1570 |
0.37 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1570 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
55 |
0.46 |
Binding ≤ 10μM |
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Functional ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
1570 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
873 |
0.39 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
55 |
0.46 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
1570 |
0.37 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
873 |
0.39 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1400 |
0.37 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
4603 |
0.34 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
1570 |
0.37 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
1570 |
0.37 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
3872 |
0.34 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
1570 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
55 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
1.20226443 |
0.57 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
6.61 |
-41.71 |
2 |
5 |
1 |
52 |
372.283 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
587541
-
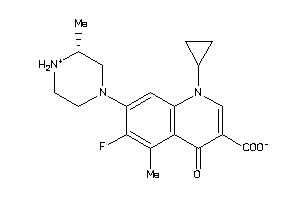
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4300 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
10.27 |
-99.09 |
2 |
6 |
0 |
82 |
359.401 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.42 |
8.46 |
-83.99 |
3 |
6 |
1 |
85 |
360.409 |
2 |
↓
|
|
|
|
Analogs
-
967787
-
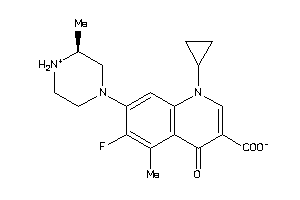
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4300 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
10.27 |
-104.27 |
2 |
6 |
0 |
82 |
359.401 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.42 |
8.43 |
-82.75 |
3 |
6 |
1 |
85 |
360.409 |
2 |
↓
|
|
|
|
Analogs
-
3929426
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2900 |
0.22 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.23 |
Binding ≤ 10μM
|
|
CP2J2-1-E |
Cytochrome P450 2J2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.20 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
94 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.25 |
Functional ≤ 10μM |
|
KCNH2-2-E |
HERG (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
204 |
0.27 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.21 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4950 |
0.21 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
128.824955 |
0.28 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
295 |
0.26 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
2340 |
0.23 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
2340 |
0.23 |
Binding ≤ 10μM
|
|
CP2J2_HUMAN |
P51589
|
Cytochrome P450 2J2, Human |
8100 |
0.20 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
128.824955 |
0.28 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
295 |
0.26 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
2900 |
0.22 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
204 |
0.27 |
Functional ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
563 |
0.25 |
Functional ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
3960 |
0.22 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
1100 |
0.24 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1995.26231 |
0.23 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
320 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.17 |
14.49 |
-42.25 |
3 |
3 |
1 |
45 |
472.693 |
9 |
↓
|
|
|
|
Analogs
-
38141475
-
-
8214628
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
11.13 |
-18.22 |
1 |
10 |
0 |
103 |
616.833 |
35 |
↓
|
|
|
|
|