|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.06 |
3.29 |
-15.59 |
6 |
9 |
0 |
149 |
678.693 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1900 |
1.33 |
Binding ≤ 10μM |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5813 |
1.22 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
1900 |
1.33 |
Binding ≤ 10μM
|
|
TRPA1_RAT |
Q6RI86
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Rat |
5813 |
1.22 |
Functional ≤ 10μM |
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
1120 |
1.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
3.02 |
-27.34 |
1 |
1 |
1 |
14 |
100.166 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.25 |
2.45 |
-1.96 |
0 |
1 |
0 |
12 |
99.158 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
5.48 |
-12.6 |
1 |
4 |
0 |
59 |
262.268 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1080 |
0.76 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4090 |
0.69 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
470 |
0.81 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4720 |
0.68 |
Binding ≤ 10μM |
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4450 |
0.68 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3210 |
0.70 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
0.14 |
-48.46 |
2 |
4 |
-1 |
81 |
153.113 |
1 |
↓
|
|
|
|
Analogs
-
13779018
-
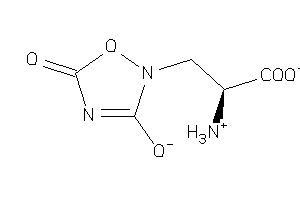
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.04 |
-4.73 |
-52.73 |
3 |
8 |
-1 |
139 |
188.119 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.04 |
-5.08 |
-89.4 |
2 |
8 |
-2 |
137 |
187.111 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.04 |
-7.92 |
-16.73 |
4 |
8 |
0 |
132 |
189.127 |
3 |
↓
|
|
|
|
Analogs
-
22066252
-
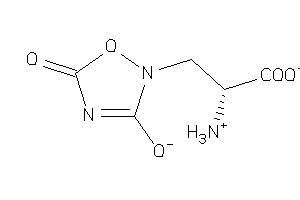
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FOLH1-1-E |
Glutamate Carboxypeptidase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.54 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
171 |
0.73 |
Binding ≤ 10μM
|
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1670 |
0.62 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM8-2-E |
Metabotropic Glutamate Receptor 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T35-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T43-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T47-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
XCT-1-E |
Cystine/glutamate Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.57 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.64 |
Functional ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
55 |
0.78 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7300 |
0.55 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
131.825674 |
0.74 |
Binding ≤ 1μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
GRIA1_MOUSE |
P23818
|
Glutamate Receptor Ionotropic, AMPA 1, Mouse |
80 |
0.76 |
Binding ≤ 1μM
|
|
GRIA2_MOUSE |
P23819
|
Glutamate Receptor Ionotropic, AMPA 2, Mouse |
80 |
0.76 |
Binding ≤ 1μM
|
|
GRIA3_MOUSE |
Q9Z2W9
|
Glutamate Receptor Ionotropic, AMPA 3, Mouse |
80 |
0.76 |
Binding ≤ 1μM
|
|
GRIA4_MOUSE |
Q9Z2W8
|
Glutamate Receptor Ionotropic, AMPA 4, Mouse |
80 |
0.76 |
Binding ≤ 1μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
200 |
0.72 |
Binding ≤ 1μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
10 |
0.86 |
Binding ≤ 1μM
|
|
GRM1_MOUSE |
P97772
|
Metabotropic Glutamate Receptor 1, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
Q80T43_MOUSE |
Q80T43
|
Metabotropic Glutamate Receptor 2, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
GRM3_MOUSE |
Q9QYS2
|
Metabotropic Glutamate Receptor 3, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
Q80T47_MOUSE |
Q80T47
|
Metabotropic Glutamate Receptor 5, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
150 |
0.73 |
Binding ≤ 1μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
200 |
0.72 |
Binding ≤ 1μM
|
|
Q80T35_MOUSE |
Q80T35
|
Metabotropic Glutamate Receptor 6, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
GRM7_MOUSE |
Q68ED2
|
Metabotropic Glutamate Receptor 7, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
GRM8_MOUSE |
P47743
|
Metabotropic Glutamate Receptor 8, Mouse |
25 |
0.82 |
Binding ≤ 1μM
|
|
XCT_HUMAN |
Q9UPY5
|
Cystine/glutamate Transporter, Human |
5000 |
0.57 |
Binding ≤ 10μM
|
|
FOLH1_HUMAN |
Q04609
|
Glutamate Carboxypeptidase II, Human |
9500 |
0.54 |
Binding ≤ 10μM
|
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
131.825674 |
0.74 |
Binding ≤ 10μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
1380.38426 |
0.63 |
Binding ≤ 10μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1_MOUSE |
P23818
|
Glutamate Receptor Ionotropic, AMPA 1, Mouse |
1300 |
0.63 |
Binding ≤ 10μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
1300 |
0.63 |
Binding ≤ 10μM
|
|
GRIA2_MOUSE |
P23819
|
Glutamate Receptor Ionotropic, AMPA 2, Mouse |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
1300 |
0.63 |
Binding ≤ 10μM
|
|
GRIA3_MOUSE |
Q9Z2W9
|
Glutamate Receptor Ionotropic, AMPA 3, Mouse |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
1300 |
0.63 |
Binding ≤ 10μM
|
|
GRIA4_MOUSE |
Q9Z2W8
|
Glutamate Receptor Ionotropic, AMPA 4, Mouse |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRM1_MOUSE |
P97772
|
Metabotropic Glutamate Receptor 1, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
10 |
0.86 |
Binding ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
200 |
0.72 |
Binding ≤ 10μM
|
|
Q80T43_MOUSE |
Q80T43
|
Metabotropic Glutamate Receptor 2, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM3_MOUSE |
Q9QYS2
|
Metabotropic Glutamate Receptor 3, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T47_MOUSE |
Q80T47
|
Metabotropic Glutamate Receptor 5, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
200 |
0.72 |
Binding ≤ 10μM
|
|
Q80T35_MOUSE |
Q80T35
|
Metabotropic Glutamate Receptor 6, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM7_MOUSE |
Q68ED2
|
Metabotropic Glutamate Receptor 7, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM8_MOUSE |
P47743
|
Metabotropic Glutamate Receptor 8, Mouse |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
1100 |
0.64 |
Functional ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
900 |
0.65 |
Functional ≤ 10μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
55 |
0.78 |
Functional ≤ 10μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
900 |
0.65 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
100 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.04 |
-4.9 |
-43.55 |
3 |
8 |
-1 |
139 |
188.119 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.04 |
-5.21 |
-87.71 |
2 |
8 |
-2 |
137 |
187.111 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.04 |
-7.81 |
-15.85 |
4 |
8 |
0 |
132 |
189.127 |
3 |
↓
|
|
|
|
Analogs
-
558
-
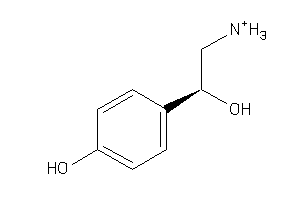
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-5-E |
Serotonin 3a (5-HT3a) Receptor (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
24 |
0.97 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.55 |
-2.59 |
-48.05 |
5 |
3 |
1 |
68 |
154.189 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.55 |
-2.98 |
-5.39 |
4 |
3 |
0 |
66 |
153.181 |
2 |
↓
|
|
|
|
Analogs
-
388198
-
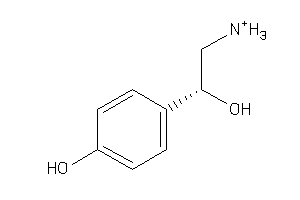
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-5-E |
Serotonin 3a (5-HT3a) Receptor (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
24 |
0.97 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.55 |
-2.67 |
-47.53 |
5 |
3 |
1 |
68 |
154.189 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.55 |
-3.12 |
-5.5 |
4 |
3 |
0 |
66 |
153.181 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
4.46 |
-50.47 |
2 |
6 |
-1 |
115 |
283.215 |
1 |
↓
|
|
|
|
Analogs
-
34618471
-
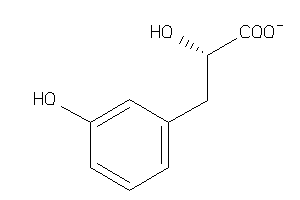
-
34618473
-
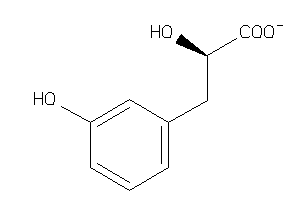
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.25 |
-4.69 |
-45.38 |
3 |
5 |
-1 |
100 |
197.166 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITAL-2-E |
Integrin Alpha-L (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
950 |
0.23 |
Binding ≤ 10μM |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80164-2-O |
HT-1080 (Fibrosarcoma Cells) (cluster #2 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
3160 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
3.89 |
-15.77 |
4 |
7 |
0 |
132 |
514.659 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-2-E |
GABA-B Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
1.44 |
Binding ≤ 10μM |
|
GABR2-2-E |
GABA-B Receptor 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
1.44 |
Binding ≤ 10μM |
|
GBRA1-7-E |
GABA Receptor Alpha-1 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
10000 |
1.00 |
Binding ≤ 10μM
|
|
GBRA2-8-E |
GABA Receptor Alpha-2 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA3-8-E |
GABA Receptor Alpha-3 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA4-7-E |
GABA Receptor Alpha-4 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA5-8-E |
GABA Receptor Alpha-5 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA6-7-E |
GABA Receptor Alpha-6 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRB1-6-E |
GABA Receptor Beta-1 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB2-5-E |
GABA Receptor Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1-6-E |
GABA Receptor Gamma-1 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG2-6-E |
GABA Receptor Gamma-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG3-7-E |
GABA Receptor Gamma-3 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRT-5-E |
GABA Receptor Theta Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
S6A11-1-E |
GABA Transporter 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
1.03 |
Binding ≤ 10μM
|
|
S6A13-1-E |
GABA Transporter 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
1.06 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
1.06 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRA1-5-E |
GABA Receptor Alpha-1 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA5-4-E |
GABA Receptor Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRA6-4-E |
GABA Receptor Alpha-6 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRR1-4-E |
GABA Receptor Rho-1 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
800 |
1.22 |
Functional ≤ 10μM
|
|
GBRR2-2-E |
GABA Receptor Rho-2 Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRR3-1-E |
GABA Receptor Rho-3 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
S6A11-2-E |
GABA Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
S6A12-1-E |
Betaine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
S6A13-2-E |
GABA Transporter 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
SC6A1-1-E |
GABA Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
Z104301-3-O |
GABA-A Receptor; Anion Channel (cluster #3 Of 8), Other |
Other |
49 |
1.46 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 5), Other |
Other |
8600 |
1.01 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
70 |
1.43 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
22 |
1.53 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
33 |
1.50 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
100 |
1.40 |
Binding ≤ 1μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
17 |
1.55 |
Binding ≤ 1μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
25 |
1.52 |
Binding ≤ 1μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
17 |
1.55 |
Binding ≤ 1μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
25 |
1.52 |
Binding ≤ 1μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
10000 |
1.00 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
22 |
1.53 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
33 |
1.50 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
SC6A1_HUMAN |
P30531
|
GABA Transporter 1, Human |
5000 |
1.06 |
Binding ≤ 10μM
|
|
SC6A1_RAT |
P23978
|
GABA Transporter 1, Rat |
1620 |
1.16 |
Binding ≤ 10μM
|
|
S6A13_RAT |
P31646
|
GABA Transporter 2, Rat |
5000 |
1.06 |
Binding ≤ 10μM
|
|
S6A11_HUMAN |
P48066
|
GABA Transporter 3, Human |
7000 |
1.03 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
100 |
1.40 |
Binding ≤ 10μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
17 |
1.55 |
Binding ≤ 10μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
25 |
1.52 |
Binding ≤ 10μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
17 |
1.55 |
Binding ≤ 10μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
25 |
1.52 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
8600 |
1.01 |
Binding ≤ 10μM
|
|
S6A12_RAT |
P48056
|
Betaine Transporter, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRR1_HUMAN |
P24046
|
GABA Receptor Rho-1 Subunit, Human |
1190 |
1.18 |
Functional ≤ 10μM
|
|
GBRR2_HUMAN |
P28476
|
GABA Receptor Rho-2 Subunit, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRR3_HUMAN |
A8MPY1
|
GABA Receptor Rho-3 Subunit, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
SC6A1_RAT |
P23978
|
GABA Transporter 1, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
S6A13_RAT |
P31646
|
GABA Transporter 2, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
S6A11_RAT |
P31647
|
GABA Transporter 3, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2000 |
1.14 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.10 |
0.98 |
-62.79 |
3 |
3 |
0 |
68 |
103.121 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.54 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.54 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.54 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
14 |
0.85 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8-2-E |
Adenylate Cyclase Type VIII (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.57 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 5), Other |
Other |
9000 |
0.54 |
Binding ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
6520 |
0.56 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
0.6 |
0.99 |
Binding ≤ 1μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.99 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
14 |
0.85 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
940 |
0.65 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
150 |
0.73 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
0.6 |
0.99 |
Binding ≤ 10μM
|
|
AA1R_BOVIN |
P28190
|
Adenosine A1 Receptor, Bovin |
2430 |
0.60 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
1310 |
0.63 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.99 |
Binding ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
1310 |
0.63 |
Binding ≤ 10μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
2700 |
0.60 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
14 |
0.85 |
Binding ≤ 10μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
150 |
0.73 |
Binding ≤ 10μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
940 |
0.65 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
150 |
0.73 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
4800 |
0.57 |
Functional ≤ 10μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
9070 |
0.54 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
10000 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
3.24 |
-11.33 |
1 |
6 |
0 |
73 |
180.167 |
0 |
↓
|
|
|
|
Analogs
-
3831599
-
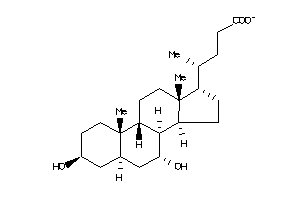
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
7.71 |
-45.97 |
2 |
4 |
-1 |
81 |
391.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.25 |
5.74 |
-7.17 |
3 |
4 |
0 |
78 |
392.58 |
4 |
↓
|
|
|
|
Analogs
-
3831599
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
7.75 |
-47.16 |
2 |
4 |
-1 |
81 |
391.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.25 |
5.77 |
-6.78 |
3 |
4 |
0 |
78 |
392.58 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.07 |
13.64 |
-44.83 |
0 |
2 |
-1 |
40 |
283.476 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
731 |
0.86 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.44 |
0.46 |
-47.41 |
4 |
2 |
1 |
48 |
138.19 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
9920 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
8800 |
0.71 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7200 |
0.72 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
7120 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
3.11 |
-51.26 |
1 |
3 |
-1 |
60 |
137.114 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.13 |
13.66 |
-43.62 |
0 |
2 |
-1 |
40 |
277.428 |
13 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.13 |
11.68 |
-5.43 |
1 |
2 |
0 |
37 |
278.436 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.20 |
-4.03 |
-29.86 |
6 |
3 |
1 |
77 |
60.08 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
930 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
802 |
0.47 |
Binding ≤ 10μM
|
|
FABPH-3-E |
Fatty Acid Binding Protein Muscle (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.43 |
Binding ≤ 10μM
|
|
FABPI-2-E |
Fatty Acid Binding Protein Intestinal (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
|
TLR2-1-E |
Toll-like Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Functional ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
5000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.06 |
2.36 |
-44.91 |
0 |
2 |
-1 |
40 |
255.422 |
14 |
↓
|
|
|
|
Analogs
-
5226647
-
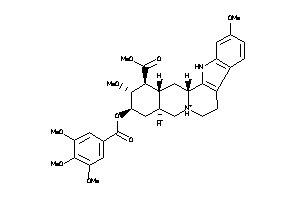
-
5226650
-
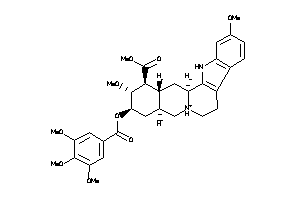
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
11.7 |
-45.84 |
2 |
11 |
1 |
119 |
609.696 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
610 |
0.36 |
ADME/T ≤ 10μM
|
|
CP2DI-1-E |
Cytochrome P450 2D18 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.34 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
94 |
0.41 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
652 |
0.36 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
81 |
0.41 |
Functional ≤ 10μM
|
|
Z50460-2-O |
Leishmania Major (cluster #2 Of 4), Other |
Other |
9530 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.18 |
-40.05 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
7.62 |
-89.58 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH4-6-E |
Carbonic Anhydrase IV (cluster #6 Of 16), Eukaryotic |
Eukaryotes |
99 |
0.75 |
Binding ≤ 10μM
|
|
AROQ-1-B |
3-dehydroquinate Dehydratase (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.98 |
3.22 |
-217.8 |
1 |
7 |
-3 |
141 |
189.099 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.98 |
1.24 |
-131.87 |
2 |
7 |
-2 |
138 |
190.107 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.98 |
-0.74 |
-64.58 |
3 |
7 |
-1 |
135 |
191.115 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.20 |
-0.91 |
-39.4 |
5 |
5 |
0 |
108 |
197.19 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
1.18 |
Binding ≤ 10μM
|
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1200 |
1.04 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
77 |
1.24 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
75 |
1.25 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
45 |
1.29 |
Functional ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
1.08 |
Functional ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
88 |
1.23 |
Functional ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
1.01 |
Functional ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
980 |
1.05 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
62 |
1.26 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
13 |
1.38 |
Binding ≤ 1μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
463 |
1.11 |
Binding ≤ 1μM
|
|
HRH3_MOUSE |
P58406
|
Histamine H3 Receptor, Mouse |
68 |
1.25 |
Binding ≤ 1μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
10 |
1.40 |
Binding ≤ 1μM
|
|
HRH4_MOUSE |
Q91ZY2
|
Histamine H4 Receptor, Mouse |
79.4328235 |
1.24 |
Binding ≤ 1μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
63.0957344 |
1.26 |
Binding ≤ 1μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.6 |
1.39 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
13 |
1.38 |
Binding ≤ 10μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
1200 |
1.04 |
Binding ≤ 10μM
|
|
HRH2_CAVPO |
P47747
|
Histamine H2 Receptor, Guinea Pig |
1200 |
1.04 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
10 |
1.40 |
Binding ≤ 10μM
|
|
HRH3_MOUSE |
P58406
|
Histamine H3 Receptor, Mouse |
68 |
1.25 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.6 |
1.39 |
Binding ≤ 10μM
|
|
HRH4_MOUSE |
Q91ZY2
|
Histamine H4 Receptor, Mouse |
79.4328235 |
1.24 |
Binding ≤ 10μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
63.0957344 |
1.26 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
1000 |
1.05 |
Functional ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1000 |
1.05 |
Functional ≤ 10μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
650 |
1.08 |
Functional ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
88 |
1.23 |
Functional ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
62 |
1.26 |
Functional ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
1800 |
1.01 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
62 |
1.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
0.85 |
-42.34 |
4 |
3 |
1 |
56 |
112.156 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.85 |
1.31 |
-96.9 |
5 |
3 |
2 |
58 |
113.164 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
-7.79 |
-55.22 |
5 |
9 |
-1 |
167 |
353.303 |
5 |
↓
|
|
|
|
Analogs
-
3919243
-
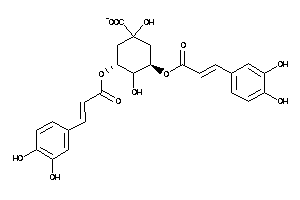
-
3947475
-
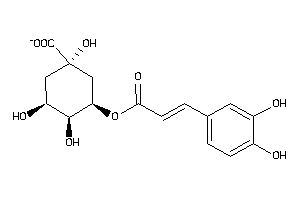
-
3947476
-
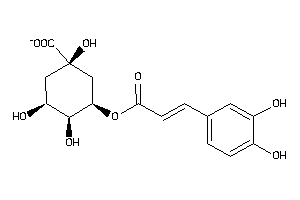
-
4096248
-
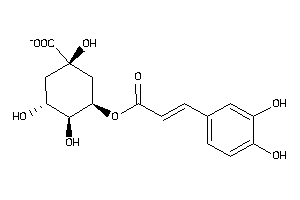
-
4102407
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
-8.03 |
-50.97 |
5 |
9 |
-1 |
167 |
353.303 |
5 |
↓
|
|
|
|
Analogs
-
3947476
-

-
4096248
-

-
4102407
-

-
6482465
-
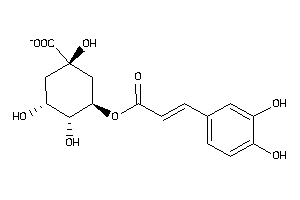
-
12153524
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
-8.39 |
-51.55 |
5 |
9 |
-1 |
167 |
353.303 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
270 |
1.02 |
Binding ≤ 10μM
|
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
29 |
1.17 |
Functional ≤ 10μM |
|
Z50588-6-O |
Canis Familiaris (cluster #6 Of 7), Other |
Other |
2700 |
0.87 |
Functional ≤ 10μM
|
|
Z50597-8-O |
Rattus Norvegicus (cluster #8 Of 12), Other |
Other |
960 |
0.94 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
3.29 |
-45.83 |
0 |
3 |
-1 |
53 |
122.103 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.27 |
3.74 |
-46.83 |
1 |
3 |
0 |
54 |
123.111 |
1 |
↓
|
|
|
|
Analogs
-
33822153
-
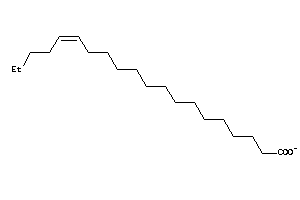
-
4529321
-
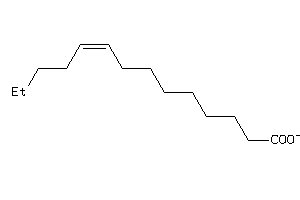
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
185 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
248 |
0.46 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4_HUMAN |
P15090
|
Fatty Acid Binding Protein Adipocyte, Human |
185 |
0.47 |
Binding ≤ 1μM
|
|
FABP5_HUMAN |
Q01469
|
Fatty Acid Binding Protein Epidermal, Human |
248 |
0.46 |
Binding ≤ 1μM
|
|
FABPL_HUMAN |
P07148
|
Fatty Acid-binding Protein, Liver, Human |
200 |
0.47 |
Binding ≤ 1μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
180 |
0.47 |
Binding ≤ 1μM
|
|
FAAH1_HUMAN |
O00519
|
Anandamide Amidohydrolase, Human |
6000 |
0.37 |
Binding ≤ 10μM
|
|
FABP4_HUMAN |
P15090
|
Fatty Acid Binding Protein Adipocyte, Human |
185 |
0.47 |
Binding ≤ 10μM
|
|
FABP5_HUMAN |
Q01469
|
Fatty Acid Binding Protein Epidermal, Human |
248 |
0.46 |
Binding ≤ 10μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
180 |
0.47 |
Binding ≤ 10μM
|
|
FABPL_HUMAN |
P07148
|
Fatty Acid-binding Protein, Liver, Human |
200 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.58 |
13.34 |
-45.08 |
0 |
2 |
-1 |
40 |
281.46 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-5-E |
Adenosine Receptor A3 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
16 |
1.36 |
Binding ≤ 10μM
|
|
GBRA1-5-E |
GABA Receptor Alpha-1 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRA2-7-E |
GABA Receptor Alpha-2 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRA3-6-E |
GABA Receptor Alpha-3 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRA4-6-E |
GABA Receptor Alpha-4 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRA5-5-E |
GABA Receptor Alpha-5 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRA6-5-E |
GABA Receptor Alpha-6 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
2 |
1.52 |
Binding ≤ 10μM
|
|
GBRB1-5-E |
GABA Receptor Beta-1 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRB2-3-E |
GABA Receptor Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRB3-3-E |
GABA Receptor Beta-3 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRD-2-E |
GABA Receptor Delta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRE-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG1-3-E |
GABA Receptor Gamma-1 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG2-4-E |
GABA Receptor Gamma-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG3-6-E |
GABA Receptor Gamma-3 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRP-2-E |
GABA Receptor Pi Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRT-3-E |
GABA Receptor Theta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA1-3-E |
GABA Receptor Alpha-1 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA2-3-E |
GABA Receptor Alpha-2 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA3-3-E |
GABA Receptor Alpha-3 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA4-3-E |
GABA Receptor Alpha-4 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRR1-2-E |
GABA Receptor Rho-1 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2300 |
0.99 |
Functional ≤ 10μM
|
|
GBRR2-1-E |
GABA Receptor Rho-2 Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
1.02 |
Functional ≤ 10μM
|
|
Z104301-6-O |
GABA-A Receptor; Anion Channel (cluster #6 Of 8), Other |
Other |
6 |
1.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
16 |
1.36 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
6 |
1.44 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
11 |
1.39 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10 |
1.40 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
16 |
1.36 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
6 |
1.44 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
11 |
1.39 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10 |
1.40 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
135 |
1.20 |
Functional ≤ 10μM
|
|
GBRR1_HUMAN |
P24046
|
GABA Receptor Rho-1 Subunit, Human |
1400 |
1.02 |
Functional ≤ 10μM
|
|
GBRR2_HUMAN |
P28476
|
GABA Receptor Rho-2 Subunit, Human |
1400 |
1.02 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.25 |
-3.98 |
-69.85 |
3 |
4 |
0 |
77 |
114.104 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.25 |
-4.38 |
-47.41 |
2 |
4 |
-1 |
75 |
113.096 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
7400 |
1.20 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
1280 |
1.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.54 |
-1.56 |
-97.63 |
6 |
2 |
2 |
55 |
90.17 |
3 |
↓
|
|
|
|
Analogs
-
39089
-
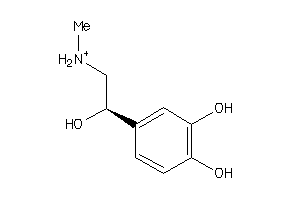
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.79 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.79 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.79 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.06 |
-2.87 |
-46.53 |
5 |
4 |
1 |
77 |
184.215 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.06 |
-4.37 |
-7.91 |
4 |
4 |
0 |
73 |
183.207 |
3 |
↓
|
|
|
|
Analogs
-
39090
-
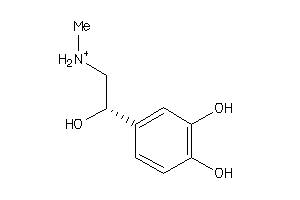
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.80 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.80 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.80 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.61 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.66 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Functional ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.71 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.71 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.71 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
2800 |
0.60 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 1), Other |
Other |
4073 |
0.58 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
400 |
0.69 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
200 |
0.72 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 1μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
630 |
0.67 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
660 |
0.67 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1600 |
0.62 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2000 |
0.61 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.84 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
13 |
0.85 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
5 |
0.89 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
3000 |
0.59 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1100 |
0.64 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
630 |
0.67 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1100 |
0.64 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
4000 |
0.58 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
4073 |
0.58 |
Functional ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
400 |
0.69 |
Functional ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
300 |
0.70 |
Functional ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
15 |
0.84 |
Functional ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
230 |
0.71 |
Functional ≤ 10μM
|
|
ADRB2_RAT |
P10608
|
Beta-2 Adrenergic Receptor, Rat |
230 |
0.71 |
Functional ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
15 |
0.84 |
Functional ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
230 |
0.71 |
Functional ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
31 |
0.81 |
Functional ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
230 |
0.71 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
400 |
0.69 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.06 |
-2.8 |
-45.1 |
5 |
4 |
1 |
77 |
184.215 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.06 |
-4.26 |
-7.35 |
4 |
4 |
0 |
73 |
183.207 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.62 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
750 |
0.71 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.83 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
750 |
0.71 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.65 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.70 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
7300 |
0.60 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 1), Other |
Other |
200 |
0.78 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1500 |
0.68 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1000 |
0.70 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
110 |
0.81 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
30 |
0.88 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
30 |
0.88 |
Binding ≤ 1μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
400 |
0.75 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.70 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.70 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.70 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1000 |
0.70 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
1500 |
0.68 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
110 |
0.81 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
30 |
0.88 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
16 |
0.91 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
30 |
0.88 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
400 |
0.75 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.70 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
3980 |
0.63 |
Binding ≤ 10μM
|
|
ADRB2_BOVIN |
Q28044
|
Beta-2 Adrenergic Receptor, Bovin |
6000 |
0.61 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.70 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.70 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.2 |
1.04 |
Functional ≤ 10μM
|
|
ADA1A_RABIT |
O02824
|
Alpha-1a Adrenergic Receptor, Rabit |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADA1D_RABIT |
O02666
|
Alpha-1d Adrenergic Receptor, Rabit |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
10000 |
0.58 |
Functional ≤ 10μM
|
|
ADRB2_RAT |
P10608
|
Beta-2 Adrenergic Receptor, Rat |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
213 |
0.78 |
Functional ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1580 |
0.68 |
Functional ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
2600 |
0.65 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
1500 |
0.68 |
Functional ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
0.1 |
1.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
-4.68 |
-50.09 |
6 |
4 |
1 |
88 |
170.188 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.04 |
-5.06 |
-7.63 |
5 |
4 |
0 |
87 |
169.18 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.60 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
-4.76 |
-51.58 |
6 |
4 |
1 |
88 |
170.188 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.04 |
-5.21 |
-8.21 |
5 |
4 |
0 |
87 |
169.18 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FYN-5-E |
Tyrosine-protein Kinase FYN (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
900 |
2.12 |
Binding ≤ 10μM
|
|
LCK-4-E |
Tyrosine-protein Kinase LCK (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
640 |
2.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
1.16 |
-44.05 |
0 |
2 |
-1 |
40 |
59.044 |
0 |
↓
|
|