|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.48 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.35 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
179 |
0.43 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
179 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1950 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
1.53 |
-8.82 |
1 |
5 |
0 |
56 |
299.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
-0.15 |
-10.09 |
1 |
5 |
0 |
56 |
388.265 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.46 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.46 |
Binding ≤ 10μM
|
|
F16P1-1-E |
Fructose-1,6-bisphosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK12-2-E |
MAP Kinase P38 Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK14-3-E |
MAP Kinase P38 Alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
630 |
0.39 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1489 |
0.37 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM |
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
143 |
0.44 |
Functional ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.47 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
211 |
0.42 |
Functional ≤ 10μM
|
|
Z100731-1-O |
Bcap37 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
4400 |
0.34 |
Functional ≤ 10μM
|
|
Z80037-1-O |
Balb/MK (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4700 |
0.34 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
820 |
0.39 |
Functional ≤ 10μM
|
|
Z80852-3-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
8310 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
0.66 |
0.58 |
Binding ≤ 1μM
|
|
F16P1_HUMAN |
P09467
|
Fructose-1,6-bisphosphatase, Human |
1000 |
0.38 |
Binding ≤ 1μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
630 |
0.39 |
Binding ≤ 1μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
70 |
0.46 |
Binding ≤ 1μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
30 |
0.48 |
Binding ≤ 1μM
|
|
EPHB2_HUMAN |
P29323
|
Ephrin Type-B Receptor 2, Human |
8100 |
0.32 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
0.66 |
0.58 |
Binding ≤ 10μM
|
|
F16P1_HUMAN |
P09467
|
Fructose-1,6-bisphosphatase, Human |
1000 |
0.38 |
Binding ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
630 |
0.39 |
Binding ≤ 10μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
70 |
0.46 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
30 |
0.48 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
8100 |
0.32 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
1400 |
0.37 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
1489 |
0.37 |
Binding ≤ 10μM
|
|
Z80852 |
Z80852
|
A-431 (Epidermoid Carcinoma Cells) |
1520 |
0.37 |
Functional ≤ 10μM
|
|
Z103205 |
Z103205
|
A431 |
4400 |
0.34 |
Functional ≤ 10μM
|
|
Z80037 |
Z80037
|
Balb/MK |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z100731 |
Z100731
|
Bcap37 (Breast Carcinoma Cells) |
8900 |
0.32 |
Functional ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
2.6 |
0.55 |
Functional ≤ 10μM
|
|
Z100492 |
Z100492
|
MCF10 (Normal Breast Epithelial Cells) |
211 |
0.42 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
1613 |
0.37 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
8900 |
0.32 |
Functional ≤ 10μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
143 |
0.44 |
Functional ≤ 10μM
|
|
ERBB4_HUMAN |
Q15303
|
Receptor Protein-tyrosine Kinase ErbB-4, Human |
49 |
0.47 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
820 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.59 |
-10.58 |
1 |
5 |
0 |
56 |
360.211 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
489 |
0.38 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
489 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.84 |
-9.82 |
1 |
5 |
0 |
56 |
313.332 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.70 |
8.31 |
-35.71 |
2 |
5 |
1 |
58 |
314.34 |
5 |
↓
|
|
|
|
Analogs
-
4554508
-
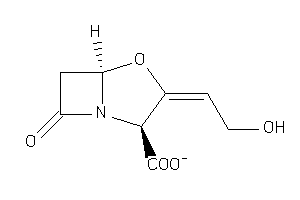
-
4554509
-
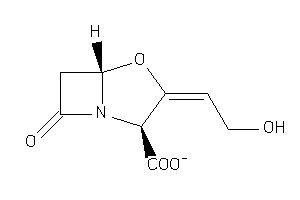
-
4554510
-
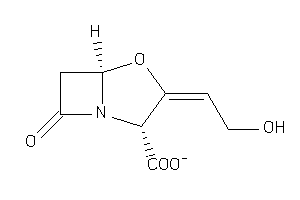
-
4554511
-
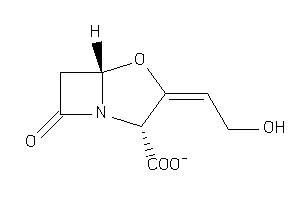
-
1169
-
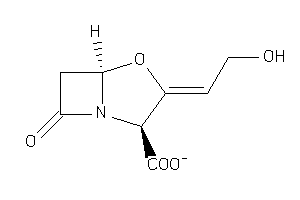
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A0ZX81-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
1720 |
0.58 |
Binding ≤ 10μM
|
|
A1E3K9-1-B |
Beta-lactamase SCO-1 (cluster #1 Of 1), Bacterial |
Bacteria |
70 |
0.72 |
Binding ≤ 10μM
|
|
A4KCT8-1-B |
Gil1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
A8RR46-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
85 |
0.71 |
Binding ≤ 10μM
|
|
AMPC-5-B |
Beta-lactamase (cluster #5 Of 6), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLA1-1-B |
Beta-lactamase L1 (cluster #1 Of 3), Bacterial |
Bacteria |
170 |
0.68 |
Binding ≤ 10μM
|
|
BLAC-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
80 |
0.71 |
Binding ≤ 10μM
|
|
BLAT-3-B |
Beta-lactamase TEM (cluster #3 Of 4), Bacterial |
Bacteria |
800 |
0.61 |
Binding ≤ 10μM
|
|
BLO1-1-B |
Beta-lactamase OXA-1 (cluster #1 Of 1), Bacterial |
Bacteria |
3200 |
0.55 |
Binding ≤ 10μM
|
|
D1MIX9-1-B |
Beta-lactamase GES-13 (cluster #1 Of 1), Bacterial |
Bacteria |
100 |
0.70 |
Binding ≤ 10μM
|
|
Q2XPY6-1-B |
Extended-spectrum Beta-lactamase CTX-M-53 (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.80 |
Binding ≤ 10μM
|
|
Q4TVR4-1-B |
Beta-lactamase SHV-55 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.77 |
Binding ≤ 10μM
|
|
Q50H31-1-B |
Class D Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
2000 |
0.57 |
Binding ≤ 10μM
|
|
Q6GWS8-1-B |
Beta-lactamase TEM-125 (cluster #1 Of 1), Bacterial |
Bacteria |
8600 |
0.51 |
Binding ≤ 10μM
|
|
Q9EXV5-1-B |
Beta-lactamase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.80 |
Binding ≤ 10μM
|
|
BLAT-1-B |
Beta-lactamase TEM (cluster #1 Of 1), Bacterial |
Bacteria |
60 |
0.72 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
40 |
0.74 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8RR46_BACCS |
A8RR46
|
Beta-lactamase, Baccs |
85 |
0.71 |
Binding ≤ 1μM
|
|
Q9EXV5_ECOLX |
Q9EXV5
|
Beta-lactamase, Ecolx |
9 |
0.80 |
Binding ≤ 1μM
|
|
AMPC_PSEAE |
P24735
|
Beta-lactamase, Pseae |
800 |
0.61 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
15 |
0.78 |
Binding ≤ 1μM
|
|
BLAC_STAAU |
P00807
|
Beta-lactamase, Staau |
80 |
0.71 |
Binding ≤ 1μM
|
|
D1MIX9_PSEAI |
D1MIX9
|
Beta-lactamase GES-13, Pseai |
100 |
0.70 |
Binding ≤ 1μM
|
|
A1E3K9_ECOLX |
A1E3K9
|
Beta-lactamase SCO-1, Ecolx |
70 |
0.72 |
Binding ≤ 1μM
|
|
BLA1_KLEPN |
P0AD64
|
Beta-lactamase SHV-1, Klepn |
170 |
0.68 |
Binding ≤ 1μM
|
|
BLA1_ECOLX |
P0AD63
|
Beta-lactamase SHV-1, Ecolx |
28 |
0.76 |
Binding ≤ 1μM
|
|
Q4TVR4_KLEPN |
Q4TVR4
|
Beta-lactamase SHV-55, Klepn |
20 |
0.77 |
Binding ≤ 1μM
|
|
BLAT_ECOLX |
P62593
|
Beta-lactamase TEM, Ecolx |
170 |
0.68 |
Binding ≤ 1μM
|
|
Q2XPY6_SALET |
Q2XPY6
|
Extended-spectrum Beta-lactamase CTX-M-53, Salet |
10 |
0.80 |
Binding ≤ 1μM
|
|
A4KCT8_9ENTR |
A4KCT8
|
Gil1, 9entr |
9 |
0.80 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
15 |
0.78 |
Binding ≤ 10μM
|
|
Q9EXV5_ECOLX |
Q9EXV5
|
Beta-lactamase, Ecolx |
9 |
0.80 |
Binding ≤ 10μM
|
|
BLAC_STAAU |
P00807
|
Beta-lactamase, Staau |
80 |
0.71 |
Binding ≤ 10μM
|
|
A8RR46_BACCS |
A8RR46
|
Beta-lactamase, Baccs |
85 |
0.71 |
Binding ≤ 10μM
|
|
A0ZX81_KLEPN |
A0ZX81
|
Beta-lactamase, Klepn |
1720 |
0.58 |
Binding ≤ 10μM
|
|
AMPC_PSEAE |
P24735
|
Beta-lactamase, Pseae |
800 |
0.61 |
Binding ≤ 10μM
|
|
D1MIX9_PSEAI |
D1MIX9
|
Beta-lactamase GES-13, Pseai |
100 |
0.70 |
Binding ≤ 10μM
|
|
BLO1_ECOLX |
P13661
|
Beta-lactamase OXA-1, Ecolx |
3200 |
0.55 |
Binding ≤ 10μM
|
|
A1E3K9_ECOLX |
A1E3K9
|
Beta-lactamase SCO-1, Ecolx |
70 |
0.72 |
Binding ≤ 10μM
|
|
BLA1_KLEPN |
P0AD64
|
Beta-lactamase SHV-1, Klepn |
170 |
0.68 |
Binding ≤ 10μM
|
|
BLA1_ECOLX |
P0AD63
|
Beta-lactamase SHV-1, Ecolx |
28 |
0.76 |
Binding ≤ 10μM
|
|
Q4TVR4_KLEPN |
Q4TVR4
|
Beta-lactamase SHV-55, Klepn |
20 |
0.77 |
Binding ≤ 10μM
|
|
BLAT_ECOLX |
P62593
|
Beta-lactamase TEM, Ecolx |
170 |
0.68 |
Binding ≤ 10μM
|
|
Q6GWS8_ECOLX |
Q6GWS8
|
Beta-lactamase TEM-125, Ecolx |
8600 |
0.51 |
Binding ≤ 10μM
|
|
Q50H31_BRAPL |
Q50H31
|
Class D Beta-lactamase, Brapl |
2000 |
0.57 |
Binding ≤ 10μM
|
|
Q2XPY6_SALET |
Q2XPY6
|
Extended-spectrum Beta-lactamase CTX-M-53, Salet |
10 |
0.80 |
Binding ≤ 10μM
|
|
A4KCT8_9ENTR |
A4KCT8
|
Gil1, 9entr |
9 |
0.80 |
Binding ≤ 10μM
|
|
BLAT_ECOLX |
P62593
|
Beta-lactamase TEM, Ecolx |
60 |
0.72 |
Functional ≤ 10μM
|
|
Z50212 |
Z50212
|
Escherichia Coli |
40 |
0.74 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.42 |
-3.38 |
-49.41 |
1 |
6 |
-1 |
90 |
198.154 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4_HUMAN |
P27487
|
Dipeptidyl Peptidase IV, Human |
7000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
7.12 |
-37.96 |
2 |
3 |
1 |
37 |
263.405 |
3 |
↓
|
|
|
|
Analogs
-
6398
-
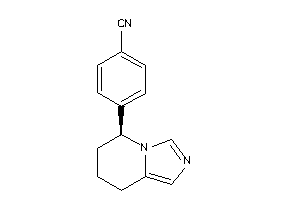
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.66 |
Binding ≤ 10μM
|
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.61 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
171 |
0.56 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
52 |
0.60 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Binding ≤ 10μM
|
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Functional ≤ 10μM
|
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.51 |
Functional ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.62 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
600 |
0.51 |
Functional ≤ 10μM
|
|
Z50740-1-O |
Cricetinae Gen. Sp. (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
9.77 |
-36.47 |
1 |
3 |
1 |
43 |
224.287 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
16 |
0.30 |
Binding ≤ 10μM
|
|
S29A1-1-E |
Equilibrative Nucleoside Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4954 |
0.21 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S29A1_HUMAN |
Q99808
|
Equilibrative Nucleoside Transporter 1, Human |
279.9 |
0.25 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
15.8489319 |
0.30 |
Binding ≤ 1μM
|
|
SCN1A_RAT |
P04774
|
Sodium Channel Protein Type I Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 1μM
|
|
SCN2A_RAT |
P04775
|
Sodium Channel Protein Type II Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 1μM
|
|
SCN3A_RAT |
P08104
|
Sodium Channel Protein Type III Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 1μM
|
|
SCN8A_RAT |
O88420
|
Sodium Channel Protein Type VIII Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 1μM
|
|
S29A1_HUMAN |
Q99808
|
Equilibrative Nucleoside Transporter 1, Human |
279.9 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
15.8489319 |
0.30 |
Binding ≤ 10μM
|
|
SCN1A_RAT |
P04774
|
Sodium Channel Protein Type I Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A_RAT |
P04775
|
Sodium Channel Protein Type II Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN3A_RAT |
P08104
|
Sodium Channel Protein Type III Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN8A_RAT |
O88420
|
Sodium Channel Protein Type VIII Alpha Subunit, Rat |
77 |
0.28 |
Binding ≤ 10μM
|
|
SCN2A_RAT |
P04775
|
Sodium Channel Protein Type II Alpha Subunit, Rat |
2400 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.33 |
15.19 |
-49.33 |
2 |
4 |
1 |
37 |
492.634 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.33 |
12.89 |
-12.84 |
1 |
4 |
0 |
36 |
491.626 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.13 |
8.03 |
-14.25 |
1 |
4 |
0 |
45 |
231.299 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
4.42 |
-23.48 |
1 |
9 |
0 |
121 |
314.257 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.93 |
1.83 |
-52.95 |
0 |
9 |
-1 |
127 |
313.249 |
4 |
↓
|
|
|
|
Analogs
-
11680893
-
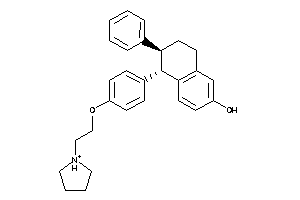
-
11680896
-
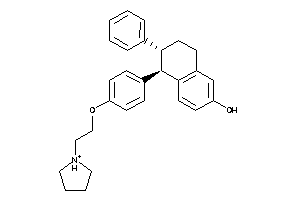
-
537882
-
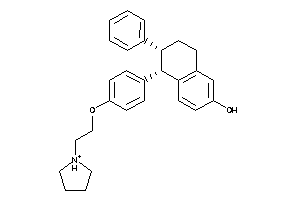
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
11 |
0.36 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
2 |
0.39 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
11.1 |
0.36 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
1.3 |
0.40 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1.8 |
0.39 |
Binding ≤ 1μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
11.1 |
0.36 |
Binding ≤ 1μM
|
|
ESR1_RAT |
P06211
|
Estrogen Receptor Alpha, Rat |
11.1 |
0.36 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
1.3 |
0.40 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1.8 |
0.39 |
Binding ≤ 10μM
|
|
ESR2_RAT |
Q62986
|
Estrogen Receptor Beta, Rat |
11.1 |
0.36 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
0.1 |
0.45 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
0.1 |
0.45 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
0.1 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
0.19 |
-37.23 |
2 |
3 |
1 |
33 |
414.569 |
6 |
↓
|
|
|
|
Analogs
-
537882
-

-
3918428
-
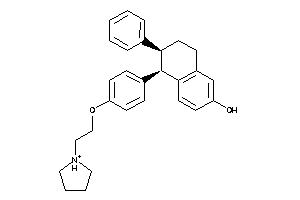
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
0.28 |
-36.68 |
2 |
3 |
1 |
34 |
414.569 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
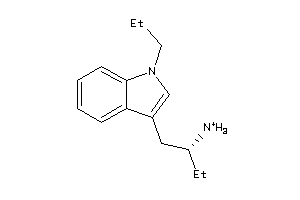
-
44407283
-
-
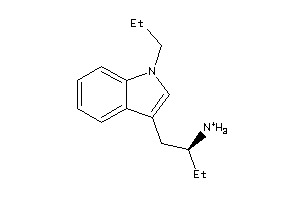
44407286
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2400 |
0.44 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
8.87 |
-44.51 |
3 |
2 |
1 |
33 |
245.39 |
6 |
↓
|
|
|
|
Analogs
-
44407283
-

-
44407286
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2400 |
0.44 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
8.94 |
-39.39 |
3 |
2 |
1 |
33 |
245.39 |
6 |
↓
|
|
|
|
Analogs
-
57278
-
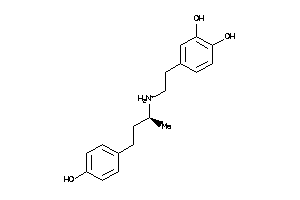
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
390 |
0.41 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
480 |
0.40 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
730 |
0.39 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
890 |
0.39 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
9820 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
9530 |
0.32 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
150 |
0.43 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
3.7 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.79 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
-
3911
-
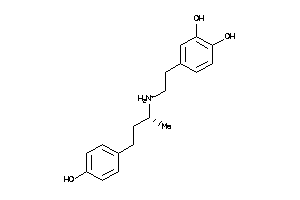
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
390 |
0.41 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
480 |
0.40 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
730 |
0.39 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
890 |
0.39 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
9820 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
9530 |
0.32 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
150 |
0.43 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
3.7 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.72 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
12.3 |
-25.65 |
3 |
3 |
1 |
41 |
304.417 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.98 |
-15.71 |
-34.21 |
9 |
8 |
1 |
158 |
268.286 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.98 |
-15.06 |
-35.68 |
9 |
8 |
1 |
158 |
268.286 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.12 |
-2.4 |
-165.36 |
3 |
9 |
-2 |
163 |
270.074 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.12 |
-1.74 |
-366.18 |
1 |
9 |
-4 |
164 |
268.058 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.12 |
-3.45 |
-71.58 |
4 |
9 |
-1 |
160 |
271.082 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CSK2B-1-E |
Casein Kinase II Beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
DYR1A-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
7.23 |
-133.03 |
0 |
5 |
-2 |
80 |
290.278 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.20 |
6.12 |
-69.86 |
1 |
5 |
-1 |
78 |
291.286 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FPPS-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.58 |
Binding ≤ 10μM
|
|
Q0GKD7-1-E |
Farnesyl Pyrophosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
|
Q197X6-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.59 |
Binding ≤ 10μM
|
|
Q8WS26-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
|
Z50418-6-O |
Trypanosoma Brucei (cluster #6 Of 6), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-17-O |
Plasmodium Falciparum (cluster #17 Of 22), Other |
Other |
1400 |
0.48 |
Functional ≤ 10μM
|
|
Z50459-5-O |
Leishmania Donovani (cluster #5 Of 8), Other |
Other |
2300 |
0.46 |
Functional ≤ 10μM
|
|
Z50472-3-O |
Toxoplasma Gondii (cluster #3 Of 4), Other |
Other |
490 |
0.52 |
Functional ≤ 10μM
|
|
Z50594-2-O |
Mus Musculus (cluster #2 Of 9), Other |
Other |
300 |
0.54 |
Functional ≤ 10μM
|
|
Z50725-4-O |
Trypanosoma Brucei Rhodesiense (cluster #4 Of 7), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50759-2-O |
Dictyostelium Discoideum (cluster #2 Of 2), Other |
Other |
3000 |
0.45 |
Functional ≤ 10μM
|
|
Z80954-2-O |
HFF (Foreskin Fibroblasts) (cluster #2 Of 4), Other |
Other |
2400 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
-3.55 |
-231.08 |
2 |
8 |
-3 |
157 |
280.089 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.23 |
-2.31 |
-381.96 |
1 |
8 |
-4 |
159 |
279.081 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.23 |
-4.53 |
-107.38 |
3 |
8 |
-2 |
154 |
281.097 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
396 |
0.53 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
119 |
0.57 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
119 |
0.57 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
249 |
0.54 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
520 |
0.52 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
119 |
0.57 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
119 |
0.57 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1140 |
0.49 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
249 |
0.54 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
2200 |
0.47 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
9.88 |
-41.85 |
1 |
2 |
1 |
28 |
229.347 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3B-3-E |
Serotonin 3b (5-HT3b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
116 |
0.88 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.67 |
1.5 |
-32.73 |
5 |
4 |
1 |
79 |
171.249 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.54 |
1.45 |
-33.39 |
5 |
4 |
1 |
77 |
171.249 |
3 |
↓
|
|
|
|
Analogs
-
5157081
-
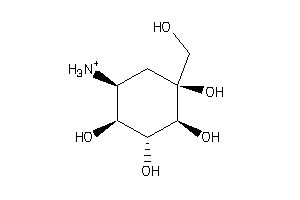
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3S,4R,5R)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol
(1R,2S,3S,4R,5R)-5-amino-1-(hydr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-10.86 |
-37.53 |
8 |
6 |
1 |
129 |
194.207 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3S,4R,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol
(1R,2S,3S,4R,5S)-5-amino-1-(hydr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-11.71 |
-41.66 |
8 |
6 |
1 |
129 |
194.207 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3S,4S,5R)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol
(1R,2S,3S,4S,5R)-5-amino-1-(hydr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-10.09 |
-46.87 |
8 |
6 |
1 |
129 |
194.207 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-10.47 |
-6.02 |
7 |
6 |
0 |
127 |
193.199 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,2S,3S,4S,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol
(1R,2S,3S,4S,5S)-5-amino-1-(hydr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.61 |
-10.55 |
-43.17 |
8 |
6 |
1 |
129 |
194.207 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.61 |
-11.15 |
-5.8 |
7 |
6 |
0 |
127 |
193.199 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104290-4-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #4 Of 4), Other |
Other |
1300 |
0.55 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
1300 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
3.79 |
-37.59 |
1 |
4 |
1 |
39 |
228.703 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.04 |
1.24 |
-5.87 |
0 |
4 |
0 |
38 |
227.695 |
4 |
↓
|
|
|
|
Analogs
-
34026476
-
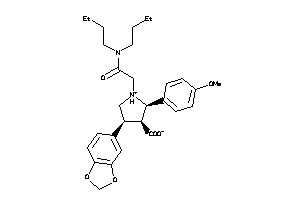
-
34026477
-
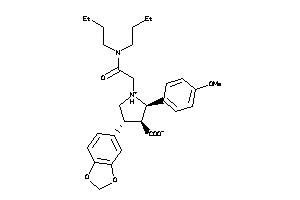
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA_RAT |
P26684
|
Endothelin Receptor ET-A, Rat |
0.1 |
0.38 |
Binding ≤ 1μM
|
|
EDNRA_HUMAN |
P25101
|
Endothelin Receptor ET-A, Human |
0.1 |
0.38 |
Binding ≤ 1μM
|
|
EDNRB_PIG |
P35463
|
Endothelin Receptor ET-B, Pig |
110 |
0.26 |
Binding ≤ 1μM
|
|
EDNRB_HUMAN |
P24530
|
Endothelin Receptor ET-B, Human |
515 |
0.24 |
Binding ≤ 1μM
|
|
EDNRA_HUMAN |
P25101
|
Endothelin Receptor ET-A, Human |
0.1 |
0.38 |
Binding ≤ 10μM
|
|
EDNRA_RAT |
P26684
|
Endothelin Receptor ET-A, Rat |
0.1 |
0.38 |
Binding ≤ 10μM
|
|
EDNRB_HUMAN |
P24530
|
Endothelin Receptor ET-B, Human |
515 |
0.24 |
Binding ≤ 10μM
|
|
EDNRB_PIG |
P35463
|
Endothelin Receptor ET-B, Pig |
110 |
0.26 |
Binding ≤ 10μM
|
|
EDNRA_RAT |
P26684
|
Endothelin Receptor ET-A, Rat |
0.1 |
0.38 |
Functional ≤ 10μM
|
|
EDNRB_RAT |
P21451
|
Endothelin Receptor ET-B, Rat |
0.1 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
1.68 |
-51.98 |
1 |
8 |
0 |
92 |
510.631 |
12 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-1-E |
Adaptor-associated Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.24 |
Binding ≤ 10μM
|
|
AKT3-2-E |
Serine/threonine-protein Kinase AKT3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.22 |
Binding ≤ 10μM
|
|
BMP2K-1-E |
BMP-2-inducible Protein Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.20 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.25 |
Binding ≤ 10μM
|
|
CLK1-1-E |
Dual Specificty Protein Kinase CLK1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
910 |
0.24 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.26 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8300 |
0.20 |
Binding ≤ 10μM
|
|
CSK21-1-E |
Casein Kinase II Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3600 |
0.22 |
Binding ≤ 10μM
|
|
CSK22-1-E |
Casein Kinase II Alpha (prime) (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.22 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
DAPK1-1-E |
Death-associated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
970 |
0.24 |
Binding ≤ 10μM
|
|
DAPK2-1-E |
Death-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM
|
|
DMPK-1-E |
Myotonin-protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.26 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5400 |
0.21 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.21 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.24 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.22 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.26 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5400 |
0.21 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.22 |
Binding ≤ 10μM
|
|
KCC1A-1-E |
CaM Kinase I Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC1D-1-E |
CaM Kinase I Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.20 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC2D-1-E |
CaM Kinase II Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.20 |
Binding ≤ 10μM
|
|
KCC4-1-E |
CaM Kinase IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KGP2-1-E |
CGMP-dependent Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.22 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
920 |
0.24 |
Binding ≤ 10μM
|
|
KKCC1-1-E |
CaM-kinase Kinase Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
Binding ≤ 10μM
|
|
KKCC2-1-E |
CaM-kinase Kinase Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.34 |
Binding ≤ 10μM
|
|
KPCD2-1-E |
Serine/threonine-protein Kinase D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.24 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.25 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
280 |
0.26 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.22 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.21 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
LATS1-1-E |
Serine/threonine-protein Kinase LATS1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.25 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.23 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.20 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.20 |
Binding ≤ 10μM
|
|
MARK2-1-E |
MAP/microtubule Affinity-regulating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.28 |
Binding ≤ 10μM
|
|
MP2K4-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
930 |
0.24 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.26 |
Binding ≤ 10μM
|
|
NUAK1-1-E |
NUAK Family SNF1-like Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.23 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.26 |
Binding ≤ 10μM
|
|
PDPK1-1-E |
3-phosphoinositide Dependent Protein Kinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.21 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.22 |
Binding ≤ 10μM
|
|
PI42B-1-E |
Phosphatidylinositol-5-phosphate 4-kinase Type-2 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
830 |
0.24 |
Binding ≤ 10μM
|
|
PI51A-1-E |
Phosphatidylinositol-4-phosphate 5-kinase Type-1 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.25 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.29 |
Binding ≤ 10μM
|
|
PIM2-1-E |
Serine/threonine-protein Kinase PIM2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.23 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.32 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.26 |
Binding ≤ 10μM
|
|
PKN2-1-E |
Protein Kinase N2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.23 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.21 |
Binding ≤ 10μM
|
|
RIOK1-1-E |
Serine/threonine-protein Kinase RIO1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.24 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.24 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.26 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.28 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.22 |
Binding ≤ 10μM
|
|
STK4-1-E |
Serine/threonine-protein Kinase MST1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.27 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.21 |
Binding ≤ 10μM
|
|
Z100405-1-O |
MV4-11 (Myeloid Leukemia Cells) (cluster #1 Of 1), Other |
Other |
1500 |
0.23 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDPK1_HUMAN |
O15530
|
3-phosphoinositide Dependent Protein Kinase-1, Human |
700 |
0.25 |
Binding ≤ 1μM
|
|
AAK1_HUMAN |
Q2M2I8
|
Adaptor-associated Kinase, Human |
900 |
0.24 |
Binding ≤ 1μM
|
|
BMP2K_HUMAN |
Q9NSY1
|
BMP-2-inducible Protein Kinase, Human |
430 |
0.25 |
Binding ≤ 1μM
|
|
DAPK1_HUMAN |
P53355
|
Death-associated Protein Kinase 1, Human |
970 |
0.24 |
Binding ≤ 1μM
|
|
MP2K4_HUMAN |
P45985
|
Dual Specificity Mitogen-activated Protein Kinase Kinase 4, Human |
930 |
0.24 |
Binding ≤ 1μM
|
|
CLK2_HUMAN |
P49760
|
Dual Specificity Protein Kinase CLK2, Human |
420 |
0.26 |
Binding ≤ 1μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
910 |
0.24 |
Binding ≤ 1μM
|
|
MK15_HUMAN |
Q8TD08
|
Mitogen-activated Protein Kinase 15, Human |
88 |
0.28 |
Binding ≤ 1μM
|
|
MYLK_HUMAN |
Q15746
|
Myosin Light Chain Kinase, Smooth Muscle, Human |
290 |
0.26 |
Binding ≤ 1μM
|
|
DMPK_HUMAN |
Q09013
|
Myotonin-protein Kinase, Human |
280 |
0.26 |
Binding ≤ 1μM
|
|
NUAK2_HUMAN |
Q9H093
|
NUAK Family SNF1-like Kinase 2, Human |
320 |
0.26 |
Binding ≤ 1μM
|
|
PI51A_HUMAN |
Q99755
|
Phosphatidylinositol-4-phosphate 5-kinase Type-1 Alpha, Human |
690 |
0.25 |
Binding ≤ 1μM
|
|
PI42B_HUMAN |
P78356
|
Phosphatidylinositol-5-phosphate 4-kinase Type-2 Beta, Human |
830 |
0.24 |
Binding ≤ 1μM
|
|
KPCA_RAT |
P05696
|
Protein Kinase C Alpha, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCA_HUMAN |
P17252
|
Protein Kinase C Alpha, Human |
360 |
0.26 |
Binding ≤ 1μM
|
|
KPCB_HUMAN |
P05771
|
Protein Kinase C Beta, Human |
4.7 |
0.33 |
Binding ≤ 1μM
|
|
KPCB_RAT |
P68403
|
Protein Kinase C Beta, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCD_RAT |
P09215
|
Protein Kinase C Delta, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
250 |
0.26 |
Binding ≤ 1μM
|
|
KPCE_HUMAN |
Q02156
|
Protein Kinase C Epsilon, Human |
600 |
0.25 |
Binding ≤ 1μM
|
|
KPCE_RAT |
P09216
|
Protein Kinase C Epsilon, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
52 |
0.29 |
Binding ≤ 1μM
|
|
KPCL_RAT |
Q64617
|
Protein Kinase C Eta, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCG_HUMAN |
P05129
|
Protein Kinase C Gamma, Human |
300 |
0.26 |
Binding ≤ 1μM
|
|
KPCG_RAT |
P63319
|
Protein Kinase C Gamma, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCT_RAT |
Q9WTQ0
|
Protein Kinase C Theta, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
KPCT_HUMAN |
Q04759
|
Protein Kinase C Theta, Human |
2.5 |
0.34 |
Binding ≤ 1μM
|
|
KPCZ_RAT |
P09217
|
Protein Kinase C Zeta, Rat |
320 |
0.26 |
Binding ≤ 1μM
|
|
PKN1_HUMAN |
Q16512
|
Protein Kinase N1, Human |
350 |
0.26 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
KS6A2_HUMAN |
Q15349
|
Ribosomal Protein S6 Kinase Alpha 2, Human |
280 |
0.26 |
Binding ≤ 1μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
280 |
0.26 |
Binding ≤ 1μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
CHK1_HUMAN |
O14757
|
Serine/threonine-protein Kinase Chk1, Human |
540 |
0.25 |
Binding ≤ 1μM
|
|
KPCD2_HUMAN |
Q9BZL6
|
Serine/threonine-protein Kinase D2, Human |
890 |
0.24 |
Binding ≤ 1μM
|
|
LATS2_HUMAN |
Q9NRM7
|
Serine/threonine-protein Kinase LATS2, Human |
640 |
0.25 |
Binding ≤ 1μM
|
|
STK4_HUMAN |
Q13043
|
Serine/threonine-protein Kinase MST1, Human |
220 |
0.27 |
Binding ≤ 1μM
|
|
STK3_HUMAN |
Q13188
|
Serine/threonine-protein Kinase MST2, Human |
960 |
0.24 |
Binding ≤ 1μM
|
|
STK3_MOUSE |
Q9JI10
|
Serine/threonine-protein Kinase MST2, Mouse |
90 |
0.28 |
Binding ≤ 1μM
|
|
PIM1_HUMAN |
P11309
|
Serine/threonine-protein Kinase PIM1, Human |
200 |
0.27 |
Binding ≤ 1μM
|
|
PIM2_HUMAN |
Q9P1W9
|
Serine/threonine-protein Kinase PIM2, Human |
130 |
0.28 |
Binding ≤ 1μM
|
|
PIM3_HUMAN |
Q86V86
|
Serine/threonine-protein Kinase PIM3, Human |
12 |
0.32 |
Binding ≤ 1μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
370 |
0.26 |
Binding ≤ 1μM
|
|
RIOK1_HUMAN |
Q9BRS2
|
Serine/threonine-protein Kinase RIO1, Human |
810 |
0.24 |
Binding ≤ 1μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
380 |
0.26 |
Binding ≤ 1μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
420 |
0.26 |
Binding ≤ 1μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
130 |
0.28 |
Binding ≤ 1μM
|
|
PDPK1_HUMAN |
O15530
|
3-phosphoinositide Dependent Protein Kinase-1, Human |
700 |
0.25 |
Binding ≤ 10μM
|
|
AAK1_HUMAN |
Q2M2I8
|
Adaptor-associated Kinase, Human |
900 |
0.24 |
Binding ≤ 10μM
|
|
BMP2K_HUMAN |
Q9NSY1
|
BMP-2-inducible Protein Kinase, Human |
2500 |
0.22 |
Binding ≤ 10μM
|
|
KCC1A_HUMAN |
Q14012
|
CaM Kinase I Alpha, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC1D_HUMAN |
Q8IU85
|
CaM Kinase I Delta, Human |
2600 |
0.22 |
Binding ≤ 10μM
|
|
KCC2A_HUMAN |
Q9UQM7
|
CaM Kinase II Alpha, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC2B_HUMAN |
Q13554
|
CaM Kinase II Beta, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC2D_HUMAN |
Q13557
|
CaM Kinase II Delta, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC2G_HUMAN |
Q13555
|
CaM Kinase II Gamma, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KCC4_HUMAN |
Q16566
|
CaM Kinase IV, Human |
6200 |
0.21 |
Binding ≤ 10μM
|
|
KKCC1_HUMAN |
Q8N5S9
|
CaM-kinase Kinase Alpha, Human |
1500 |
0.23 |
Binding ≤ 10μM
|
|
KKCC2_HUMAN |
Q96RR4
|
CaM-kinase Kinase Beta, Human |
1100 |
0.24 |
Binding ≤ 10μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
5400 |
0.21 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3200 |
0.22 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
3600 |
0.22 |
Binding ≤ 10μM
|
|
CSK22_HUMAN |
P19784
|
Casein Kinase II Alpha (prime), Human |
3400 |
0.22 |
Binding ≤ 10μM
|
|
KGP2_HUMAN |
Q13237
|
CGMP-dependent Protein Kinase 2, Human |
3100 |
0.22 |
Binding ≤ 10μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
1400 |
0.23 |
Binding ≤ 10μM
|
|
DAPK1_HUMAN |
P53355
|
Death-associated Protein Kinase 1, Human |
970 |
0.24 |
Binding ≤ 10μM
|
|
DAPK2_HUMAN |
Q9UIK4
|
Death-associated Protein Kinase 2, Human |
2600 |
0.22 |
Binding ≤ 10μM
|
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
1300 |
0.24 |
Binding ≤ 10μM
|
|
MP2K4_HUMAN |
P45985
|
Dual Specificity Mitogen-activated Protein Kinase Kinase 4, Human |
930 |
0.24 |
Binding ≤ 10μM
|
|
CLK2_HUMAN |
P49760
|
Dual Specificity Protein Kinase CLK2, Human |
420 |
0.26 |
Binding ≤ 10μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
1900 |
0.23 |
Binding ≤ 10μM
|
|
DYR1B_HUMAN |
Q9Y463
|
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B, Human |
5400 |
0.21 |
Binding ≤ 10μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
910 |
0.24 |
Binding ≤ 10μM
|
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
5100 |
0.21 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
2600 |
0.22 |
Binding ≤ 10μM
|
|
MARK2_HUMAN |
Q7KZI7
|
MAP/microtubule Affinity-regulating Kinase 2, Human |
2300 |
0.23 |
Binding ≤ 10μM
|
|
MK15_HUMAN |
Q8TD08
|
Mitogen-activated Protein Kinase 15, Human |
88 |
0.28 |
Binding ≤ 10μM
|
|
M3K9_HUMAN |
P80192
|
Mitogen-activated Protein Kinase Kinase Kinase 9, Human |
1400 |
0.23 |
Binding ≤ 10μM
|
|
M4K1_HUMAN |
Q92918
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1, Human |
2600 |
0.22 |
Binding ≤ 10μM
|
|
M4K3_HUMAN |
Q8IVH8
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3, Human |
8400 |
0.20 |
Binding ≤ 10μM
|
|
M4K5_HUMAN |
Q9Y4K4
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5, Human |
8400 |
0.20 |
Binding ≤ 10μM
|
|
MYLK_HUMAN |
Q15746
|
Myosin Light Chain Kinase, Smooth Muscle, Human |
290 |
0.26 |
Binding ≤ 10μM
|
|
DMPK_HUMAN |
Q09013
|
Myotonin-protein Kinase, Human |
280 |
0.26 |
Binding ≤ 10μM
|
|
NUAK1_HUMAN |
O60285
|
NUAK Family SNF1-like Kinase 1, Human |
1800 |
0.23 |
Binding ≤ 10μM
|
|
NUAK2_HUMAN |
Q9H093
|
NUAK Family SNF1-like Kinase 2, Human |
320 |
0.26 |
Binding ≤ 10μM
|
|
PI51A_HUMAN |
Q99755
|
Phosphatidylinositol-4-phosphate 5-kinase Type-1 Alpha, Human |
690 |
0.25 |
Binding ≤ 10μM
|
|
PI42B_HUMAN |
P78356
|
Phosphatidylinositol-5-phosphate 4-kinase Type-2 Beta, Human |
830 |
0.24 |
Binding ≤ 10μM
|
|
PHKG1_HUMAN |
Q16816
|
Phosphorylase Kinase Gamma Subunit 1, Human |
2700 |
0.22 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
7000 |
0.21 |
Binding ≤ 10μM
|
|
KPCA_RAT |
P05696
|
Protein Kinase C Alpha, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCA_HUMAN |
P17252
|
Protein Kinase C Alpha, Human |
360 |
0.26 |
Binding ≤ 10μM
|
|
KPCB_RAT |
P68403
|
Protein Kinase C Beta, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCB_HUMAN |
P05771
|
Protein Kinase C Beta, Human |
4.7 |
0.33 |
Binding ≤ 10μM
|
|
KPCD_HUMAN |
Q05655
|
Protein Kinase C Delta, Human |
250 |
0.26 |
Binding ≤ 10μM
|
|
KPCD_RAT |
P09215
|
Protein Kinase C Delta, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCE_HUMAN |
Q02156
|
Protein Kinase C Epsilon, Human |
600 |
0.25 |
Binding ≤ 10μM
|
|
KPCE_RAT |
P09216
|
Protein Kinase C Epsilon, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCL_RAT |
Q64617
|
Protein Kinase C Eta, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCL_HUMAN |
P24723
|
Protein Kinase C Eta, Human |
52 |
0.29 |
Binding ≤ 10μM
|
|
KPCG_HUMAN |
P05129
|
Protein Kinase C Gamma, Human |
300 |
0.26 |
Binding ≤ 10μM
|
|
KPCG_RAT |
P63319
|
Protein Kinase C Gamma, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCT_RAT |
Q9WTQ0
|
Protein Kinase C Theta, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
KPCT_HUMAN |
Q04759
|
Protein Kinase C Theta, Human |
2.5 |
0.34 |
Binding ≤ 10μM
|
|
KPCZ_RAT |
P09217
|
Protein Kinase C Zeta, Rat |
320 |
0.26 |
Binding ≤ 10μM
|
|
PKN1_HUMAN |
Q16512
|
Protein Kinase N1, Human |
350 |
0.26 |
Binding ≤ 10μM
|
|
PKN2_HUMAN |
Q16513
|
Protein Kinase N2, Human |
1500 |
0.23 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
KS6A2_HUMAN |
Q15349
|
Ribosomal Protein S6 Kinase Alpha 2, Human |
1500 |
0.23 |
Binding ≤ 10μM
|
|
KS6A3_HUMAN |
P51812
|
Ribosomal Protein S6 Kinase Alpha 3, Human |
2200 |
0.23 |
Binding ≤ 10μM
|
|
KS6A4_HUMAN |
O75676
|
Ribosomal Protein S6 Kinase Alpha 4, Human |
6100 |
0.21 |
Binding ≤ 10μM
|
|
KS6A5_HUMAN |
O75582
|
Ribosomal Protein S6 Kinase Alpha 5, Human |
1300 |
0.24 |
Binding ≤ 10μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
2300 |
0.23 |
Binding ≤ 10μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
1100 |
0.24 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
STK33_HUMAN |
Q9BYT3
|
Serine/threonine-protein Kinase 33, Human |
3100 |
0.22 |
Binding ≤ 10μM
|
|
AKT3_HUMAN |
Q9Y243
|
Serine/threonine-protein Kinase AKT3, Human |
1400 |
0.23 |
Binding ≤ 10μM
|
|
AURKC_HUMAN |
Q9UQB9
|
Serine/threonine-protein Kinase Aurora-C, Human |
3000 |
0.22 |
Binding ≤ 10μM
|
|
CHK1_HUMAN |
O14757
|
Serine/threonine-protein Kinase Chk1, Human |
540 |
0.25 |
Binding ≤ 10μM
|
|
KPCD2_HUMAN |
Q9BZL6
|
Serine/threonine-protein Kinase D2, Human |
890 |
0.24 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
1100 |
0.24 |
Binding ≤ 10μM
|
|
LATS1_HUMAN |
O95835
|
Serine/threonine-protein Kinase LATS1, Human |
2600 |
0.22 |
Binding ≤ 10μM
|
|
LATS2_HUMAN |
Q9NRM7
|
Serine/threonine-protein Kinase LATS2, Human |
640 |
0.25 |
Binding ≤ 10μM
|
|
STK4_HUMAN |
Q13043
|
Serine/threonine-protein Kinase MST1, Human |
220 |
0.27 |
Binding ≤ 10μM
|
|
STK3_HUMAN |
Q13188
|
Serine/threonine-protein Kinase MST2, Human |
960 |
0.24 |
Binding ≤ 10μM
|
|
STK3_MOUSE |
Q9JI10
|
Serine/threonine-protein Kinase MST2, Mouse |
90 |
0.28 |
Binding ≤ 10μM
|
|
CDK16_HUMAN |
Q00536
|
Serine/threonine-protein Kinase PCTAIRE-1, Human |
8000 |
0.20 |
Binding ≤ 10μM
|
|
PIM1_HUMAN |
P11309
|
Serine/threonine-protein Kinase PIM1, Human |
200 |
0.27 |
Binding ≤ 10μM
|
|
PIM2_HUMAN |
Q9P1W9
|
Serine/threonine-protein Kinase PIM2, Human |
130 |
0.28 |
Binding ≤ 10μM
|
|
PIM3_HUMAN |
Q86V86
|
Serine/threonine-protein Kinase PIM3, Human |
12 |
0.32 |
Binding ≤ 10μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
370 |
0.26 |
Binding ≤ 10μM
|
|
RIOK1_HUMAN |
Q9BRS2
|
Serine/threonine-protein Kinase RIO1, Human |
810 |
0.24 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
380 |
0.26 |
Binding ≤ 10μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
5800 |
0.21 |
Binding ≤ 10μM
|
|
HCK_HUMAN |
P08631
|
Tyrosine-protein Kinase HCK, Human |
2800 |
0.22 |
Binding ≤ 10μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
420 |
0.26 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
1300 |
0.24 |
Binding ≤ 10μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
130 |
0.28 |
Binding ≤ 10μM
|
|
Z100405 |
Z100405
|
MV4-11 (Myeloid Leukemia Cells) |
1500 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
0.01 |
-62.92 |
2 |
7 |
1 |
73 |
469.565 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A4-2-E |
Serotonin Transporter (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
6.36 |
-52.43 |
2 |
6 |
1 |
79 |
385.185 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.93 |
4.94 |
-6.92 |
1 |
6 |
0 |
74 |
384.177 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.54 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
430 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
7.74 |
-42.6 |
1 |
4 |
1 |
45 |
209.269 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.24 |
7.23 |
-11.3 |
0 |
4 |
0 |
44 |
208.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATS5-1-E |
ADAMTS5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.27 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.96 |
-54.51 |
1 |
5 |
-1 |
86 |
414.89 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP2_HUMAN |
P08253
|
Matrix Metalloproteinase-2, Human |
2000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
10.01 |
-45.5 |
1 |
5 |
-1 |
86 |
414.89 |
7 |
↓
|
|
|
|
Analogs
-
31555252
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
35 |
0.40 |
Binding ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
4224 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
10.42 |
-12.17 |
1 |
4 |
0 |
51 |
365.795 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.51 |
8.97 |
-41.42 |
2 |
4 |
1 |
52 |
366.803 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM |
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM |
|
Q6IE76-1-E |
Squalene Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.35 |
7.28 |
-68.22 |
6 |
7 |
0 |
124 |
470.705 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.35 |
8.42 |
-109.32 |
7 |
7 |
0 |
126 |
470.705 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.35 |
7.44 |
-75.79 |
6 |
7 |
0 |
121 |
470.705 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
390 |
0.39 |
Binding ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
86 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
8.99 |
-43.28 |
3 |
4 |
1 |
46 |
310.421 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
8.66 |
-69.9 |
3 |
7 |
0 |
112 |
440.606 |
14 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
6.41 |
-48.25 |
4 |
8 |
0 |
131 |
442.475 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
0.27 |
-63.3 |
1 |
6 |
-1 |
87 |
473.332 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
180 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
1.43 |
-36.53 |
2 |
2 |
1 |
20 |
265.38 |
4 |
↓
|
|