|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
8.03 |
-9.4 |
1 |
5 |
0 |
55 |
352.434 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1aR,4aS,7R,7aS,7bS)-1,1,7-trimethyl-4-methylene-2,3,4a,5,6,7,7a,7b-octahydro-1aH-cyclopropa[e]azule
(1aR,4aS,7R,7aS,7bS)-1,1,7-trime…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
9.27 |
-0.48 |
0 |
0 |
0 |
0 |
204.357 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
2.35 |
-11.66 |
1 |
4 |
0 |
55 |
202.213 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.48 |
2.35 |
-11.88 |
1 |
4 |
0 |
55 |
202.213 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
0.43 |
-10.02 |
3 |
6 |
0 |
96 |
302.282 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.74 |
1.42 |
-49.9 |
2 |
6 |
-1 |
99 |
301.274 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.74 |
1.09 |
-51.12 |
2 |
6 |
-1 |
99 |
301.274 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7700 |
0.34 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3400 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
9.18 |
-46.23 |
1 |
3 |
1 |
23 |
280.347 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.50 |
6.83 |
-7.69 |
0 |
3 |
0 |
22 |
279.339 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
6830 |
0.56 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
6700 |
0.56 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
6180 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
8250 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
4.26 |
-48.19 |
0 |
4 |
-1 |
59 |
181.167 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
8.79 |
-48.03 |
2 |
3 |
1 |
34 |
296.39 |
4 |
↓
|
|
|
|
Analogs
-
33831555
-
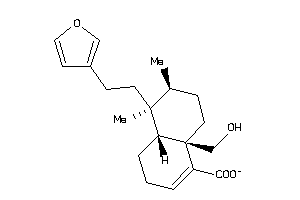
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.86 |
-53.01 |
0 |
3 |
-1 |
53 |
315.433 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
9.03 |
-7.28 |
1 |
3 |
0 |
50 |
316.441 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
6.59 |
-56.84 |
0 |
4 |
-1 |
66 |
179.151 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1080 |
0.76 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4090 |
0.69 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
470 |
0.81 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4720 |
0.68 |
Binding ≤ 10μM |
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4450 |
0.68 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3210 |
0.70 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
0.14 |
-48.46 |
2 |
4 |
-1 |
81 |
153.113 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
0.24 |
-50.82 |
2 |
4 |
-1 |
81 |
153.113 |
1 |
↓
|
|
|
|
Analogs
-
4482687
-
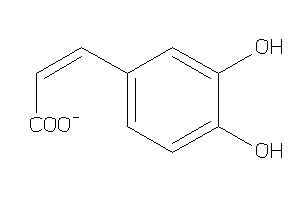
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8710 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
7870 |
0.55 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM
|
|
O49150-1-E |
5-lipoxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3060 |
0.59 |
Binding ≤ 10μM
|
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
2900 |
0.60 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.72 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O49150_SOLTU |
O49150
|
5-lipoxygenase, Soltu |
3000 |
0.59 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
7870 |
0.55 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
8710 |
0.54 |
Binding ≤ 10μM
|
|
PTN1_HUMAN |
P18031
|
Protein-tyrosine Phosphatase 1B, Human |
3060 |
0.59 |
Binding ≤ 10μM
|
|
LOX1_SOYBN |
P08170
|
Seed Lipoxygenase-1, Soybn |
3000 |
0.59 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
190 |
0.72 |
Functional ≤ 10μM
|
|
Z100766 |
Z100766
|
Radical Scavenging Activity |
2800 |
0.60 |
Functional ≤ 10μM
|
|
Z50185 |
Z50185
|
Staphylococcus Aureus |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186 |
Z50186
|
Staphylococcus Epidermidis |
2780 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.43 |
-49.59 |
2 |
4 |
-1 |
81 |
179.151 |
2 |
↓
|
|
|
|
Analogs
-
43510608
-
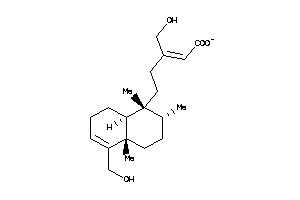
Draw
Identity
99%
90%
80%
70%
Popular Name:
(Z)-5-[(1S,2R,4aR,8aR)-5-(hydroxymethyl)-1,2,4a-trimethyl-2,3,4,7,8,8a-hexahydronaphthalen-1-yl]-3-(
(Z)-5-[(1S,2R,4aR,8aR)-5-(hydrox…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
5.33 |
-54.39 |
2 |
4 |
-1 |
81 |
335.464 |
6 |
↓
|
|
|
|
Analogs
-
4098855
-
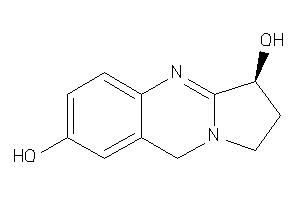
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
3.34 |
-26.34 |
2 |
3 |
1 |
37 |
189.238 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
1.04 |
3.34 |
-26.08 |
2 |
3 |
1 |
37 |
189.238 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.04 |
2.91 |
-8.05 |
1 |
3 |
0 |
36 |
188.23 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7600 |
0.48 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6300 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
4.48 |
-42.27 |
3 |
3 |
1 |
40 |
205.281 |
3 |
↓
|
|
|
|
Analogs
-
733
-
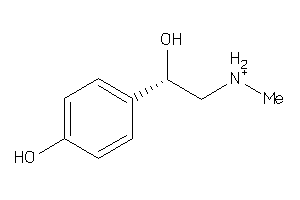
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
-0.72 |
-42.44 |
4 |
3 |
1 |
57 |
168.216 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
-2.17 |
-5.01 |
3 |
3 |
0 |
52 |
167.208 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.55 |
10.27 |
-54.71 |
2 |
4 |
-1 |
81 |
471.702 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.55 |
8.29 |
-7.67 |
3 |
4 |
0 |
78 |
472.71 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.49 |
12 |
-107.45 |
1 |
5 |
-2 |
100 |
484.677 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.49 |
10.31 |
-48.34 |
2 |
5 |
-1 |
98 |
485.685 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.49 |
8.55 |
-6.69 |
3 |
5 |
0 |
95 |
486.693 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-2-E |
Solute Carrier Family 22 Member 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6761 |
0.80 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6_MOUSE |
Q8VC69
|
Solute Carrier Family 22 Member 6, Mouse |
6700 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.15 |
2.5 |
-104.02 |
0 |
4 |
-2 |
80 |
130.099 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
930 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
802 |
0.47 |
Binding ≤ 10μM
|
|
FABPH-3-E |
Fatty Acid Binding Protein Muscle (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.43 |
Binding ≤ 10μM
|
|
FABPI-2-E |
Fatty Acid Binding Protein Intestinal (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
|
TLR2-1-E |
Toll-like Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Functional ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
5000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.06 |
2.36 |
-44.91 |
0 |
2 |
-1 |
40 |
255.422 |
14 |
↓
|
|
|
|
Analogs
-
1529229
-
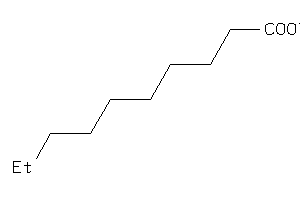
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.45 |
14.36 |
-44.86 |
0 |
2 |
-1 |
40 |
297.503 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.45 |
13.24 |
-6.37 |
1 |
2 |
0 |
37 |
298.511 |
17 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
2.72 |
-0.8 |
0 |
0 |
0 |
0 |
204.357 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
-0.44 |
-6.18 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.50 |
Binding ≤ 10μM
|
|
KCNH2-3-E |
HERG (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4898 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.06 |
4.9 |
-11.47 |
0 |
6 |
0 |
62 |
194.194 |
0 |
↓
|
|
|
|
Analogs
-
4096340
-
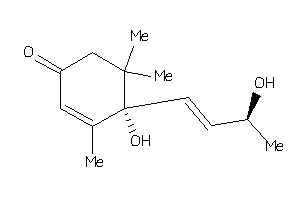
-
4095719
-
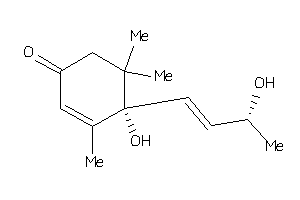
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
1.61 |
-7.75 |
2 |
3 |
0 |
58 |
224.3 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMY1-2-E |
Salivary Alpha-amylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9640 |
0.33 |
Binding ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1640 |
0.39 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
0.17 |
-23.17 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.20 |
0.76 |
-57.58 |
3 |
6 |
-1 |
114 |
285.231 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.20 |
0.98 |
-57.28 |
3 |
6 |
-1 |
114 |
285.231 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-2-B |
Beta-lactamase (cluster #2 Of 6), Bacterial |
Bacteria |
4000 |
0.34 |
Binding ≤ 10μM
|
|
AMPH-2-B |
Penicillin-binding Protein AmpH (cluster #2 Of 2), Bacterial |
Bacteria |
4000 |
0.34 |
Binding ≤ 10μM
|
|
MDH-1-B |
Malate Dehydrogenase (cluster #1 Of 1), Bacterial |
Bacteria |
6000 |
0.33 |
Binding ≤ 10μM
|
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
9800 |
0.32 |
Binding ≤ 10μM
|
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.47 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2470 |
0.36 |
Binding ≤ 10μM
|
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6990 |
0.33 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.33 |
Binding ≤ 10μM
|
|
AK1A1-1-E |
Aldehyde Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2320 |
0.36 |
Binding ≤ 10μM
|
|
AK1CL-1-E |
Aldo-keto Reductase Family 1 Member C21 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.33 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2850 |
0.35 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM
|
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
2680 |
0.35 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9390 |
0.32 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
9030 |
0.32 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5410 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-15-E |
Carbonic Anhydrase II (cluster #15 Of 15), Eukaryotic |
Eukaryotes |
2540 |
0.36 |
Binding ≤ 10μM
|
|
CAH3-6-E |
Carbonic Anhydrase III (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
7890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6810 |
0.33 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
6170 |
0.33 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4840 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.50 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
DHB2-1-E |
Estradiol 17-beta-dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1540 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2100 |
0.36 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
2100 |
0.36 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.35 |
Binding ≤ 10μM
|
|
LOX12-2-E |
Arachidonate 12-lipoxygenase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
440 |
0.40 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
500 |
0.40 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PA21B-2-E |
Phospholipase A2 Group 1B (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.47 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CB-1-E |
PI3-kinase P110-beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-1-E |
PI3-kinase P110-delta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-1-E |
PI3-kinase P110-gamma Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.40 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7100 |
0.33 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.38 |
Binding ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.39 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4097 |
0.34 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.49 |
ADME/T ≤ 10μM
|
|
Z102178-2-O |
Liver Microsomes (cluster #2 Of 2), Other |
Other |
7500 |
0.33 |
Functional ≤ 10μM
|
|
Z102342-1-O |
Liver (cluster #1 Of 1), Other |
Other |
6000 |
0.33 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8500 |
0.32 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
9600 |
0.32 |
Functional ≤ 10μM
|
|
Z81000-1-O |
HT-22 (Hippocampal Cells) (cluster #1 Of 1), Other |
Other |
2980 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
R1AB-1-V |
Replicase Polyprotein 1ab (cluster #1 Of 1), Viral |
Viruses |
8100 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
45.3 |
0.47 |
Binding ≤ 1μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
14.8 |
0.50 |
Binding ≤ 1μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1000 |
0.38 |
Binding ≤ 1μM
|
|
LOX12_HUMAN |
P18054
|
Arachidonate 12-lipoxygenase, Human |
440 |
0.40 |
Binding ≤ 1μM
|
|
LOX15_RABIT |
P12530
|
Arachidonate 15-lipoxygenase, Rabit |
430 |
0.41 |
Binding ≤ 1μM
|
|
LOX5_RAT |
P12527
|
Arachidonate 5-lipoxygenase, Rat |
200 |
0.43 |
Binding ≤ 1μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
790 |
0.39 |
Binding ≤ 1μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
850 |
0.39 |
Binding ≤ 1μM
|
|
CSK2B_HUMAN |
P67870
|
Casein Kinase II Beta, Human |
850 |
0.39 |
Binding ≤ 1μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
450 |
0.40 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
12 |
0.50 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
77 |
0.45 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
7.8 |
0.52 |
Binding ≤ 1μM
|
|
Q965D5_PLAFA |
Q965D5
|
Enoyl-acyl-carrier Protein Reductase, Plafa |
22 |
0.49 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
900 |
0.38 |
Binding ≤ 1μM
|
|
AOFA_BOVIN |
P21398
|
Monoamine Oxidase A, Bovin |
10 |
0.51 |
Binding ≤ 1μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
680 |
0.39 |
Binding ≤ 1μM
|
|
PIM1_HUMAN |
P11309
|
Serine/threonine-protein Kinase PIM1, Human |
25 |
0.48 |
Binding ≤ 1μM
|
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
5400 |
0.34 |
Binding ≤ 10μM
|
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
45.3 |
0.47 |
Binding ≤ 10μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
2470 |
0.36 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
6990 |
0.33 |
Binding ≤ 10μM
|
|
AK1A1_RAT |
P51635
|
Aldehyde Reductase, Rat |
2320 |
0.36 |
Binding ≤ 10μM
|
|
AK1CL_MOUSE |
Q91WR5
|
Aldo-keto Reductase Family 1 Member C21, Mouse |
6900 |
0.33 |
Binding ≤ 10μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
2850 |
0.35 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1000 |
0.38 |
Binding ≤ 10μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
14.8 |
0.50 |
Binding ≤ 10μM
|
|
LOX12_HUMAN |
P18054
|
Arachidonate 12-lipoxygenase, Human |
440 |
0.40 |
Binding ≤ 10μM
|
|
LOX15_HUMAN |
P16050
|
Arachidonate 15-lipoxygenase, Human |
2200 |
0.36 |
Binding ≤ 10μM
|
|
LOX15_RABIT |
P12530
|
Arachidonate 15-lipoxygenase, Rabit |
430 |
0.41 |
Binding ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
790 |
0.39 |
Binding ≤ 10μM
|
|
LOX5_RAT |
P12527
|
Arachidonate 5-lipoxygenase, Rat |
200 |
0.43 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
6900 |
0.33 |
Binding ≤ 10μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
4000 |
0.34 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2680 |
0.35 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
2540 |
0.36 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
8100 |
0.32 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
7890 |
0.32 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
6810 |
0.33 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
6170 |
0.33 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
4840 |
0.34 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
9390 |
0.32 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
9030 |
0.32 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
5410 |
0.34 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
1100 |
0.38 |
Binding ≤ 10μM
|
|
CSK2B_HUMAN |
P67870
|
Casein Kinase II Beta, Human |
1100 |
0.38 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
450 |
0.40 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
12 |
0.50 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
77 |
0.45 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
7.8 |
0.52 |
Binding ≤ 10μM
|
|
Q965D5_PLAFA |
Q965D5
|
Enoyl-acyl-carrier Protein Reductase, Plafa |
1090 |
0.38 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
900 |
0.38 |
Binding ≤ 10μM
|
|
DHB2_HUMAN |
P37059
|
Estradiol 17-beta-dehydrogenase 2, Human |
1540 |
0.37 |
Binding ≤ 10μM
|
|
Q965D7_PLAFA |
Q965D7
|
Fatty Acid Synthase, Plafa |
1500 |
0.37 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
2100 |
0.36 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
2100 |
0.36 |
Binding ≤ 10μM
|
|
LGUL_HUMAN |
Q04760
|
Glyoxalase I, Human |
3200 |
0.35 |
Binding ≤ 10μM
|
|
MDH_THETH |
P10584
|
Malate Dehydrogenase, Theth |
6000 |
0.33 |
Binding ≤ 10μM
|
|
AOFA_BOVIN |
P21398
|
Monoamine Oxidase A, Bovin |
10 |
0.51 |
Binding ≤ 10μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
2800 |
0.35 |
Binding ≤ 10μM
|
|
MRP1_HUMAN |
P33527
|
Multidrug Resistance-associated Protein 1, Human |
2400 |
0.36 |
Binding ≤ 10μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
680 |
0.39 |
Binding ≤ 10μM
|
|
AMPH_ECOLI |
P0AD70
|
Penicillin-binding Protein AmpH, Ecoli |
4000 |
0.34 |
Binding ≤ 10μM
|
|
PA21B_HUMAN |
P04054
|
Phospholipase A2 Group 1B, Human |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
P85A_HUMAN |
P27986
|
PI3-kinase P85-alpha Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
P85B_HUMAN |
O00459
|
PI3-kinase P85-beta Subunit, Human |
3800 |
0.34 |
Binding ≤ 10μM
|
|
R1AB_CVHSA |
P0C6X7
|
Replicase Polyprotein 1ab, Cvhsa |
8100 |
0.32 |
Binding ≤ 10μM
|
|
PIM1_HUMAN |
P11309
|
Serine/threonine-protein Kinase PIM1, Human |
1100 |
0.38 |
Binding ≤ 10μM
|
|
NANH_CLOPE |
P10481
|
Sialidase, Clope |
1700 |
0.37 |
Binding ≤ 10μM
|
|
TRY1_HUMAN |
P07477
|
Trypsin I, Human |
7100 |
0.33 |
Binding ≤ 10μM
|
|
XDH_BOVIN |
P80457
|
Xanthine Dehydrogenase, Bovin |
1200 |
0.38 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
2620 |
0.36 |
Binding ≤ 10μM
|
|
LOX5_RAT |
P12527
|
Arachidonate 5-lipoxygenase, Rat |
10000 |
0.32 |
Functional ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
3000 |
0.35 |
Functional ≤ 10μM
|
|
Z81000 |
Z81000
|
HT-22 (Hippocampal Cells) |
2980 |
0.35 |
Functional ≤ 10μM
|
|
Z81072 |
Z81072
|
Jurkat (Acute Leukemic T-cells) |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z102342 |
Z102342
|
Liver |
1800 |
0.37 |
Functional ≤ 10μM
|
|
Z102178 |
Z102178
|
Liver Microsomes |
7500 |
0.33 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
2511.88643 |
0.36 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
9600 |
0.32 |
Functional ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
1191 |
0.38 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
4097 |
0.34 |
ADME/T ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
23 |
0.49 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
-2.9 |
-13.58 |
5 |
7 |
0 |
131 |
302.238 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.94 |
-2.61 |
-43.8 |
4 |
7 |
-1 |
134 |
301.23 |
1 |
↓
|
|
|
|
Analogs
-
9212394
-
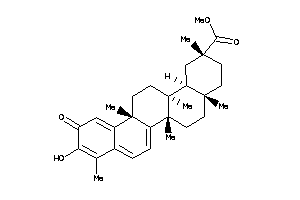
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
200 |
0.28 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
400 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.67 |
12.78 |
-16.52 |
1 |
4 |
0 |
64 |
464.646 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
6.18 |
10.75 |
-10.5 |
2 |
4 |
0 |
67 |
464.646 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
2.6 |
-13.3 |
4 |
6 |
0 |
111 |
354.358 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
1200 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
3.79 |
-14.2 |
2 |
7 |
0 |
98 |
344.319 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2162335
-
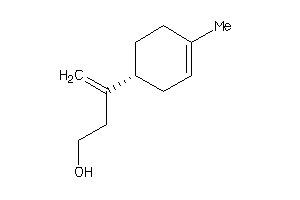
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
-0.72 |
-2.39 |
1 |
1 |
0 |
20 |
166.264 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
-0.17 |
-14.48 |
0 |
4 |
0 |
48 |
275.263 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
8000 |
0.34 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.35 |
Binding ≤ 10μM
|
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.50 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1330 |
0.39 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
47 |
0.49 |
Binding ≤ 10μM
|
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1050 |
0.40 |
Binding ≤ 10μM
|
|
DHB2-1-E |
Estradiol 17-beta-dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
360 |
0.43 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.36 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.36 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2200 |
0.38 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
0.37 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.34 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.37 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.39 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.36 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1060 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9700 |
0.33 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.41 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
716 |
0.41 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.49 |
ADME/T ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR_HUMAN |
P35869
|
Aryl Hydrocarbon Receptor, Human |
28 |
0.50 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
47 |
0.49 |
Binding ≤ 1μM
|
|
DHB2_HUMAN |
P37059
|
Estradiol 17-beta-dehydrogenase 2, Human |
360 |
0.43 |
Binding ≤ 1μM
|
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
4000 |
0.36 |
Binding ≤ 10μM
|
|
ALDR_RAT |
P07943
|
Aldose Reductase, Rat |
1330 |
0.39 |
Binding ≤ 10μM
|
|
LOX15_RABIT |
P12530
|
Arachidonate 15-lipoxygenase, Rabit |
2200 |
0.38 |
Binding ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
2700 |
0.37 |
Binding ≤ 10μM
|
|
AHR_HUMAN |
P35869
|
Aryl Hydrocarbon Receptor, Human |
28 |
0.50 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
4700 |
0.36 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
47 |
0.49 |
Binding ≤ 10μM
|
|
DHB1_HUMAN |
P14061
|
Estradiol 17-beta-dehydrogenase 1, Human |
1050 |
0.40 |
Binding ≤ 10μM
|
|
DHB2_HUMAN |
P37059
|
Estradiol 17-beta-dehydrogenase 2, Human |
360 |
0.43 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
3500 |
0.36 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
3500 |
0.36 |
Binding ≤ 10μM
|
|
MRP1_HUMAN |
P33527
|
Multidrug Resistance-associated Protein 1, Human |
2400 |
0.37 |
Binding ≤ 10μM
|
|
NOX4_HUMAN |
Q9NPH5
|
NADPH Oxidase 4, Human |
1200 |
0.39 |
Binding ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
6700 |
0.34 |
Binding ≤ 10μM
|
|
NANH_CLOPE |
P10481
|
Sialidase, Clope |
8000 |
0.34 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
1060 |
0.40 |
Binding ≤ 10μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
9700 |
0.33 |
Functional ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
632 |
0.41 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
716 |
0.41 |
ADME/T ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
43 |
0.49 |
ADME/T ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6000 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
-0.82 |
-11.52 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.43 |
-0.55 |
-41.88 |
3 |
6 |
-1 |
114 |
285.231 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
0.24 |
-9.28 |
2 |
4 |
0 |
67 |
168.148 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80901-2-O |
HaCaT (Keratinocytes) (cluster #2 Of 2), Other |
Other |
2800 |
0.52 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80901 |
Z80901
|
HaCaT (Keratinocytes) |
2800 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.35 |
-14.8 |
2 |
5 |
0 |
80 |
208.169 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.30 |
Binding ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
2230 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-2.24 |
-14.57 |
3 |
7 |
0 |
109 |
330.292 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
2010 |
0.33 |
Binding ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 3), Other |
Other |
7600 |
0.30 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
-2.43 |
-7.81 |
2 |
4 |
0 |
58 |
324.376 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
2.62 |
-12.14 |
2 |
5 |
0 |
80 |
284.267 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
2.83 |
-10.23 |
2 |
4 |
0 |
71 |
228.203 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.80 |
3.56 |
-58.09 |
1 |
4 |
-1 |
73 |
227.195 |
0 |
↓
|
|