|
|
Analogs
-
17378760
-
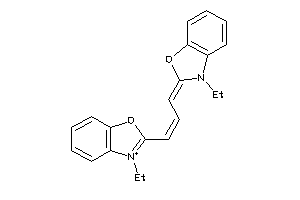
-
106856
-
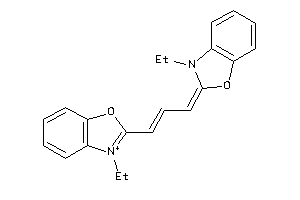
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.24 |
11.98 |
-21.63 |
0 |
4 |
1 |
35 |
333.411 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
14.41 |
-12.32 |
2 |
5 |
0 |
62 |
443.567 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-2-E |
Cannabinoid CB1 Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
480 |
0.33 |
Binding ≤ 10μM |
|
CNR2-2-E |
Cannabinoid CB2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.40 |
Functional ≤ 10μM
|
|
CNR2-2-E |
Cannabinoid CB2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.40 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
480 |
0.33 |
Binding ≤ 1μM |
|
CNR1_RAT |
P20272
|
Cannabinoid CB1 Receptor, Rat |
100 |
0.36 |
Binding ≤ 1μM
|
|
CNR2_RAT |
Q9QZN9
|
Cannabinoid CB2 Receptor, Rat |
100 |
0.36 |
Binding ≤ 1μM
|
|
FAAH1_HUMAN |
O00519
|
Anandamide Amidohydrolase, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
480 |
0.33 |
Binding ≤ 10μM |
|
CNR1_RAT |
P20272
|
Cannabinoid CB1 Receptor, Rat |
100 |
0.36 |
Binding ≤ 10μM
|
|
CNR2_RAT |
Q9QZN9
|
Cannabinoid CB2 Receptor, Rat |
100 |
0.36 |
Binding ≤ 10μM
|
|
CNR2_HUMAN |
P34972
|
Cannabinoid CB2 Receptor, Human |
1300 |
0.31 |
Binding ≤ 10μM |
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
23 |
0.40 |
Functional ≤ 10μM
|
|
CNR2_HUMAN |
P34972
|
Cannabinoid CB2 Receptor, Human |
17 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
11.89 |
-8.62 |
2 |
4 |
0 |
67 |
378.553 |
18 |
↓
|
|
|
|
Analogs
-
34223604
-
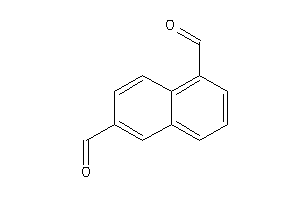
-
39263082
-
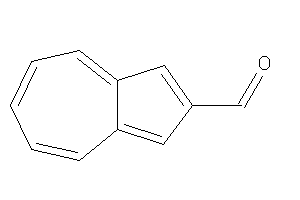
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
1.72 |
-7.64 |
0 |
1 |
0 |
17 |
156.184 |
1 |
↓
|
|
|
|
Analogs
-
34223604
-

-
39263082
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
1.71 |
-6.68 |
0 |
1 |
0 |
17 |
156.184 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
-4.06 |
-11.06 |
2 |
5 |
0 |
68 |
338.359 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HPRT-1-E |
Hypoxanthine-guanine Phosphoribosyltransferase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8980 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HPRT_HUMAN |
P00492
|
Hypoxanthine-guanine Phosphoribosyltransferase, Human |
8980 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
1.6 |
-6.78 |
0 |
0 |
0 |
0 |
228.294 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.09 |
0.24 |
-3.12 |
4 |
2 |
0 |
52 |
108.144 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
1.66 |
-6.6 |
0 |
0 |
0 |
0 |
228.294 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
2.4 |
-3.73 |
0 |
1 |
0 |
17 |
90.147 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
6289 |
0.23 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
6800 |
0.23 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.00 |
13.2 |
-76.08 |
4 |
4 |
2 |
56 |
414.597 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
1639
-
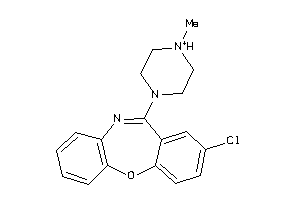
-
931
-
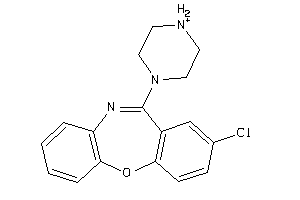
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.45 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
21 |
0.47 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
18 |
0.47 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
18 |
0.47 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
21 |
0.47 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
21 |
0.47 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
22 |
0.47 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
14 |
0.48 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6 |
0.50 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
15 |
0.48 |
Binding ≤ 1μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
43 |
0.45 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
18 |
0.47 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
21 |
0.47 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
21 |
0.47 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
22 |
0.47 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
14 |
0.48 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
5500 |
0.32 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6 |
0.50 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
15 |
0.48 |
Binding ≤ 10μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
43 |
0.45 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
7.92 |
-8.16 |
0 |
4 |
0 |
33 |
327.815 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
-0.25 |
-7.28 |
2 |
2 |
0 |
43 |
87.122 |
2 |
↓
|
|
|
|
Analogs
-
33954854
-
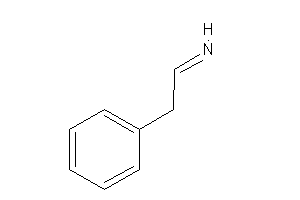
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
3.06 |
-5.22 |
1 |
2 |
0 |
33 |
135.166 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2400 |
0.72 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.42 |
-3.56 |
0 |
0 |
0 |
0 |
142.201 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
7.3 |
-3.72 |
0 |
0 |
0 |
0 |
142.201 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
E1BN64-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.53 |
Binding ≤ 10μM
|
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.46 |
Binding ≤ 10μM
|
|
PDE3A-2-E |
Phosphodiesterase 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE3B-2-E |
Phosphodiesterase 3B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4B-2-E |
Phosphodiesterase 4B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4C-2-E |
Phosphodiesterase 4C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE5A-2-E |
Phosphodiesterase 5A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.54 |
Binding ≤ 10μM
|
|
Q9XSW7-1-E |
Phosphodiesterase 3A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
843 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102213-1-O |
Blood (cluster #1 Of 2), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
122 |
0.60 |
Functional ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
9000 |
0.44 |
Functional ≤ 10μM
|
|
Z50588-6-O |
Canis Familiaris (cluster #6 Of 7), Other |
Other |
700 |
0.54 |
Functional ≤ 10μM
|
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
7700 |
0.45 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9XSW7_PIG |
Q9XSW7
|
Phosphodiesterase 3A, Pig |
843 |
0.53 |
Binding ≤ 1μM
|
|
PDE3A_RAT |
Q62865
|
Phosphodiesterase 3A, Rat |
740 |
0.54 |
Binding ≤ 1μM
|
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
150 |
0.60 |
Binding ≤ 1μM
|
|
PDE3B_RAT |
Q63085
|
Phosphodiesterase 3B, Rat |
740 |
0.54 |
Binding ≤ 1μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
150 |
0.60 |
Binding ≤ 1μM
|
|
E1BN64_BOVIN |
E1BN64
|
Phosphodiesterase 3B, Bovin |
1000 |
0.53 |
Binding ≤ 1μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
740 |
0.54 |
Binding ≤ 1μM
|
|
PDE1A_BOVIN |
P14100
|
Phosphodiesterase 1A, Bovin |
5500 |
0.46 |
Binding ≤ 10μM
|
|
PDE3A_RAT |
Q62865
|
Phosphodiesterase 3A, Rat |
740 |
0.54 |
Binding ≤ 10μM
|
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
1100 |
0.52 |
Binding ≤ 10μM
|
|
Q9XSW7_PIG |
Q9XSW7
|
Phosphodiesterase 3A, Pig |
843 |
0.53 |
Binding ≤ 10μM
|
|
PDE3B_RAT |
Q63085
|
Phosphodiesterase 3B, Rat |
740 |
0.54 |
Binding ≤ 10μM
|
|
E1BN64_BOVIN |
E1BN64
|
Phosphodiesterase 3B, Bovin |
1000 |
0.53 |
Binding ≤ 10μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
1100 |
0.52 |
Binding ≤ 10μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
10000 |
0.44 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
10000 |
0.44 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
10000 |
0.44 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
10000 |
0.44 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
10000 |
0.44 |
Binding ≤ 10μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
740 |
0.54 |
Binding ≤ 10μM
|
|
PDE5A_BOVIN |
Q28156
|
Phosphodiesterase 5A, Bovin |
5000 |
0.46 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
122 |
0.60 |
Functional ≤ 10μM
|
|
Z102213 |
Z102213
|
Blood |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50588 |
Z50588
|
Canis Familiaris |
1600 |
0.51 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
4700 |
0.47 |
Functional ≤ 10μM
|
|
PGH1_HUMAN |
P23219
|
Cyclooxygenase-1, Human |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50589 |
Z50589
|
Felis Catus |
6500 |
0.45 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
100 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
4.19 |
-16.01 |
1 |
4 |
0 |
70 |
211.224 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
2.16 |
-43.19 |
0 |
4 |
-1 |
73 |
210.216 |
1 |
↓
|
|
|
|
Analogs
-
8660419
-
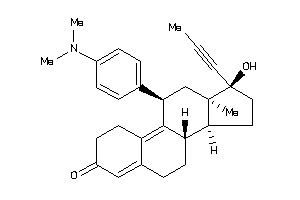
-
8681718
-
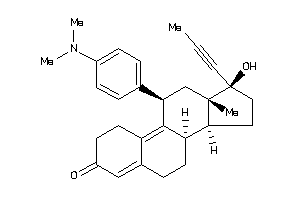
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
12.13 |
-10.13 |
1 |
3 |
0 |
41 |
429.604 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
6.34 |
10.15 |
-7.73 |
2 |
3 |
0 |
44 |
429.604 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.34 |
10.92 |
-70.79 |
3 |
3 |
0 |
45 |
430.612 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
3.82 |
-17.6 |
1 |
2 |
0 |
21 |
114.173 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
5935212
-
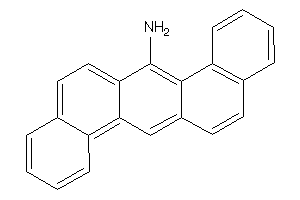
-
6118330
-
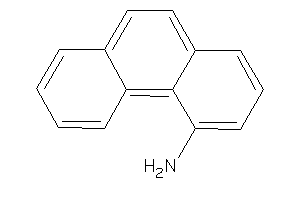
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
-0.37 |
-6.26 |
2 |
1 |
0 |
26 |
193.249 |
0 |
↓
|
|
|
|
Analogs
-
39119022
-
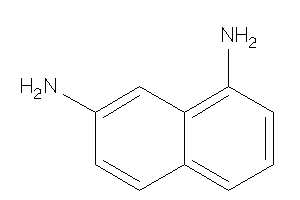
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
4.72 |
-4.41 |
2 |
1 |
0 |
26 |
143.189 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MIF-1-E |
Macrophage Migration Inhibitory Factor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.52 |
Binding ≤ 10μM
|
|
TYRO-1-E |
Tyrosinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5230 |
0.37 |
Binding ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
7100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-3.11 |
-13.08 |
3 |
5 |
0 |
90 |
270.24 |
1 |
↓
|
|
|
|
Analogs
-
4098919
-
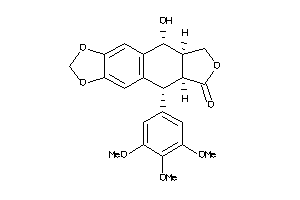
-
4098920
-
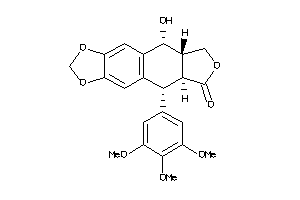
-
4475321
-
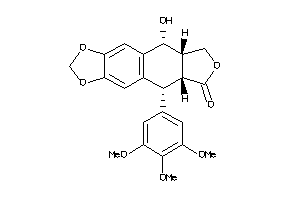
-
11682059
-
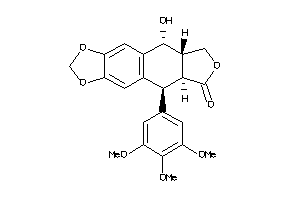
-
16969582
-
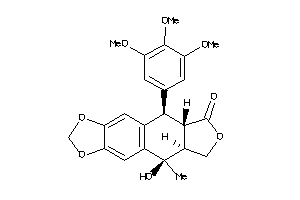
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.37 |
Binding ≤ 10μM
|
|
O46399-1-E |
Beta Tubulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q9MZB0-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Z100695-1-O |
WRL68 (Embryonic Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
4800 |
0.25 |
Functional ≤ 10μM
|
|
Z101973-1-O |
Paracentrotus Lividus (cluster #1 Of 1), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z50651-2-O |
Vesicular Stomatitis Virus (cluster #2 Of 2), Other |
Other |
100 |
0.33 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z80110-1-O |
CV-1 (Kidney Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM |
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.36 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
52 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2750 |
0.26 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
8100 |
0.24 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
30 |
0.35 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.24 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
14 |
0.37 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
460 |
0.30 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
18 |
0.36 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7630 |
0.24 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.38 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM |
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80866-1-O |
GLC4 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.34 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 4), Other |
Other |
150 |
0.32 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
16 |
0.36 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
69 |
0.33 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.28 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 1), Other |
Other |
150 |
0.32 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
34 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
23 |
0.36 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
5 |
0.39 |
ADME/T ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
9 |
0.38 |
ADME/T ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 1), Other |
Other |
31 |
0.35 |
ADME/T ≤ 10μM
|
|
VE2-1-V |
Human Papillomavirus Regulatory Protein E2 (cluster #1 Of 1), Viral |
Viruses |
1050 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR_HUMAN |
P04150
|
Glucocorticoid Receptor, Human |
14 |
0.37 |
Binding ≤ 1μM
|
|
TBA1A_PIG |
P02550
|
Tubulin Alpha Chain, Pig |
460 |
0.30 |
Binding ≤ 1μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
460 |
0.30 |
Binding ≤ 1μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB4B_HUMAN |
P68371
|
Tubulin Beta-2 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB3_HUMAN |
Q13509
|
Tubulin Beta-3 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB4A_HUMAN |
P04350
|
Tubulin Beta-4 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB8_HUMAN |
Q3ZCM7
|
Tubulin Beta-8 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
O46399_SHEEP |
O46399
|
Beta Tubulin, Sheep |
3000 |
0.26 |
Binding ≤ 10μM
|
|
GCR_HUMAN |
P04150
|
Glucocorticoid Receptor, Human |
14 |
0.37 |
Binding ≤ 10μM
|
|
VE2_HPV1A |
P03118
|
Regulatory Protein E2, Hpv1a |
1050 |
0.28 |
Binding ≤ 10μM
|
|
TBA1A_PIG |
P02550
|
Tubulin Alpha Chain, Pig |
1560 |
0.27 |
Binding ≤ 10μM
|
|
Q862F3_BOVIN |
Q862F3
|
Tubulin Alpha Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
Q9MZB0_SHEEP |
Q9MZB0
|
Tubulin Alpha-1 Chain, Sheep |
3000 |
0.26 |
Binding ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
460 |
0.30 |
Binding ≤ 10μM
|
|
Q862L2_BOVIN |
Q862L2
|
Tubulin Alpha-1 Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBA4A_HUMAN |
P68366
|
Tubulin Alpha-1 Chain, Human |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBA1A_HUMAN |
Q71U36
|
Tubulin Alpha-3 Chain, Human |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB2B_RAT |
Q3KRE8
|
Tubulin Beta Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB2B_BOVIN |
Q6B856
|
Tubulin Beta Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBB_PIG |
P02554
|
Tubulin Beta Chain, Pig |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB4B_HUMAN |
P68371
|
Tubulin Beta-2 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB3_HUMAN |
Q13509
|
Tubulin Beta-3 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB3_RAT |
Q4QRB4
|
Tubulin Beta-3 Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB4A_HUMAN |
P04350
|
Tubulin Beta-4 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB5_HUMAN |
P07437
|
Tubulin Beta-5 Chain, Human |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB8_HUMAN |
Q3ZCM7
|
Tubulin Beta-8 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBG1_RAT |
P83888
|
Tubulin Gamma-1 Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
Z80002 |
Z80002
|
1A9 (Ovarian Adenocarcinoma Cells) |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80697 |
Z80697
|
Bel-7402 (Hepatoma Cells) |
9.1 |
0.38 |
Functional ≤ 10μM
|
|
Z80054 |
Z80054
|
Caco-2 (Colon Adenocarcinoma Cells) |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
7.2 |
0.38 |
Functional ≤ 10μM
|
|
Z80110 |
Z80110
|
CV-1 (Kidney Cells) |
29 |
0.35 |
Functional ≤ 10μM
|
|
Z80121 |
Z80121
|
DMS-79 (Small Cell Lung Carcinoma Cells) |
23 |
0.36 |
Functional ≤ 10μM
|
|
Z80125 |
Z80125
|
DU-145 (Prostate Carcinoma) |
2970 |
0.26 |
Functional ≤ 10μM
|
|
Z80847 |
Z80847
|
FaDu (Pharyngeal Carcinoma Cells) |
7.2 |
0.38 |
Functional ≤ 10μM
|
|
Z80866 |
Z80866
|
GLC4 Cell Line |
150 |
0.32 |
Functional ≤ 10μM
|
|
Z81335 |
Z81335
|
HCT-15 (Colon Adenocarcinoma Cells) |
1000 |
0.28 |
Functional ≤ 10μM
|
|
Z80936 |
Z80936
|
HEK293 (Embryonic Kidney Fibroblasts) |
17 |
0.36 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
100 |
0.33 |
Functional ≤ 10μM |
|
Z81020 |
Z81020
|
HepG2 (Hepatoblastoma Cells) |
24 |
0.36 |
Functional ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
2750 |
0.26 |
Functional ≤ 10μM
|
|
Z80164 |
Z80164
|
HT-1080 (Fibrosarcoma Cells) |
8100 |
0.24 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
11 |
0.37 |
Functional ≤ 10μM
|
|
Z50602 |
Z50602
|
Human Herpesvirus 1 |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
2.9 |
0.40 |
Functional ≤ 10μM
|
|
Z81244 |
Z81244
|
J774 (Macrophage Cells) |
16.4 |
0.36 |
Functional ≤ 10μM
|
|
Z80819 |
Z80819
|
J774.2 |
11.3 |
0.37 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
30 |
0.35 |
Functional ≤ 10μM |
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
0.5 |
0.43 |
Functional ≤ 10μM |
|
Z81135 |
Z81135
|
L6 (Skeletal Muscle Myoblast Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
15 |
0.37 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
26 |
0.35 |
Functional ≤ 10μM
|
|
Z80284 |
Z80284
|
MOLT-3 (T-lymphoblastic Leukemia Cells) |
14 |
0.37 |
Functional ≤ 10μM
|
|
Z80354 |
Z80354
|
OVCAR-3 (Ovarian Adenocarcinoma Cells) |
460 |
0.30 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z101973 |
Z101973
|
Paracentrotus Lividus |
20 |
0.36 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
10 |
0.37 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80414 |
Z80414
|
Raji (B-lymphoblastic Cells) |
18 |
0.36 |
Functional ≤ 10μM
|
|
Z80427 |
Z80427
|
RKO (Colon Carcinoma) |
29 |
0.35 |
Functional ≤ 10μM
|
|
Q862F3_BOVIN |
Q862F3
|
Tubulin Alpha Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
Q862L2_BOVIN |
Q862L2
|
Tubulin Alpha-1 Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
350 |
0.30 |
Functional ≤ 10μM
|
|
TBB2B_BOVIN |
Q6B856
|
Tubulin Beta Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z50651 |
Z50651
|
Vesicular Stomatitis Virus |
100 |
0.33 |
Functional ≤ 10μM
|
|
Z100695 |
Z100695
|
WRL68 (Embryonic Hepatoma Cells) |
4800 |
0.25 |
Functional ≤ 10μM
|
|
Z80052 |
Z80052
|
C8166 (Leukemic T-cells) |
150 |
0.32 |
ADME/T ≤ 10μM
|
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
3 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135 |
Z81135
|
L6 (Skeletal Muscle Myoblast Cells) |
10 |
0.37 |
ADME/T ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
31 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
12.5 |
0.37 |
ADME/T ≤ 10μM
|
|
Z80583 |
Z80583
|
Vero (Kidney Cells) |
16 |
0.36 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4.95 |
-16.81 |
1 |
8 |
0 |
93 |
414.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
2.92 |
-3.93 |
1 |
2 |
0 |
37 |
168.236 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3831417
-
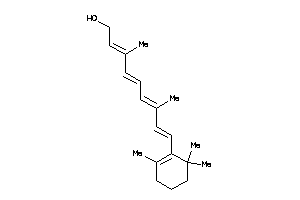
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
9.81 |
-3.68 |
1 |
1 |
0 |
20 |
286.459 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
9.07 |
-2.89 |
1 |
1 |
0 |
20 |
286.459 |
5 |
↓
|
|
|
|
Analogs
-
38220408
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
0.5 |
-11.29 |
2 |
3 |
0 |
58 |
138.122 |
1 |
↓
|
|
|
|
Analogs
-
3800039
-
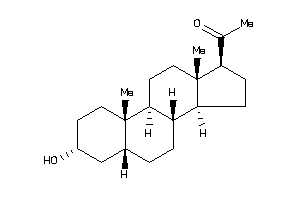
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA1-2-E |
GABA Receptor Alpha-1 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRB2-2-E |
GABA Receptor Beta-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
91 |
0.43 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
10000 |
0.30 |
Functional ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
5500 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
74 |
0.43 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
51 |
0.44 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
74 |
0.43 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
2610 |
0.34 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
160 |
0.41 |
Functional ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
160 |
0.41 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.30 |
Functional ≤ 10μM
|
|
Z50573 |
Z50573
|
Xenopus Laevis |
420 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
8.25 |
-5.76 |
1 |
2 |
0 |
37 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3651042
-
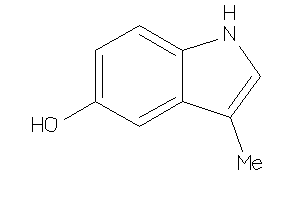
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
-1.28 |
-8.63 |
2 |
3 |
0 |
53 |
175.187 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
-2.38 |
-4.89 |
2 |
5 |
0 |
76 |
103.081 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
2.14 |
-6.64 |
1 |
2 |
0 |
29 |
68.079 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
2.68 |
-31.31 |
2 |
2 |
1 |
30 |
69.087 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
3.75 |
-4.16 |
2 |
2 |
0 |
30 |
69.087 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
2.23 |
-0.43 |
0 |
0 |
0 |
0 |
136.238 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
2.24 |
-0.43 |
0 |
0 |
0 |
0 |
136.238 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
6.02 |
-0.75 |
0 |
0 |
0 |
0 |
181.449 |
0 |
↓
|
|
|
|
Analogs
-
1577245
-
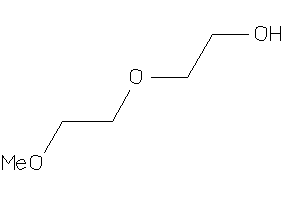
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
-2.68 |
-13.57 |
2 |
7 |
0 |
87 |
282.333 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.27 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1250 |
0.28 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
NPM-1-E |
Nucleophosmin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.27 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1660 |
0.28 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1660 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.37 |
Binding ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
2520 |
0.27 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
810 |
0.29 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
980 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
2 |
0.42 |
Binding ≤ 1μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
0.2 |
0.47 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
5 |
0.40 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
24.5 |
0.37 |
Binding ≤ 1μM
|
|
ALK_HUMAN |
Q9UM73
|
ALK Tyrosine Kinase Receptor, Human |
2500 |
0.27 |
Binding ≤ 10μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
2 |
0.42 |
Binding ≤ 10μM
|
|
FGFR1_HUMAN |
P11362
|
Fibroblast Growth Factor Receptor 1, Human |
1250 |
0.28 |
Binding ≤ 10μM
|
|
NPM_HUMAN |
P06748
|
Nucleophosmin, Human |
2500 |
0.27 |
Binding ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
1660 |
0.28 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
1660 |
0.28 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
0.2 |
0.47 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
5 |
0.40 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
24.5 |
0.37 |
Binding ≤ 10μM
|
|
Z80036 |
Z80036
|
BaF3 (IL-3-dependent Pro-B-cells) |
1800 |
0.28 |
Functional ≤ 10μM
|
|
Z80152 |
Z80152
|
HCT-8 (Ileocecal Adenocarcinoma) |
1131 |
0.29 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
1210 |
0.29 |
Functional ≤ 10μM
|
|
Z81331 |
Z81331
|
SW-620 (Colon Adenocarcinoma Cells) |
1130 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
14.64 |
-13.18 |
1 |
5 |
0 |
60 |
443.359 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1260 |
0.27 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.27 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Binding ≤ 10μM
|
|
DAPK3-1-E |
Death-associated Protein Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5700 |
0.24 |
Binding ≤ 10μM
|
|
DMPK-1-E |
Myotonin-protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.23 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.32 |
Binding ≤ 10μM
|
|
EPHA1-1-E |
Ephrin Type-A Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.24 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.28 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7300 |
0.23 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.25 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.25 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.24 |
Binding ≤ 10μM
|
|
IRAK3-1-E |
Interleukin-1 Receptor-associated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.29 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.24 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
630 |
0.28 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
990 |
0.27 |
Binding ≤ 10μM
|
|
MK04-1-E |
Mitogen-activated Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.25 |
Binding ≤ 10μM
|
|
MK06-1-E |
Mitogen-activated Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.26 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.25 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.30 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
360 |
0.29 |
Binding ≤ 10μM
|
|
MYLK2-1-E |
Myosin Light Chain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.26 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.25 |
Binding ≤ 10μM
|
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6400 |
0.23 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.24 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.26 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.28 |
Binding ≤ 10μM
|
|
SIK2-1-E |
Serine/threonine-protein Kinase SIK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.26 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
920 |
0.27 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.24 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.25 |
Binding ≤ 10μM
|
|
ST17B-1-E |
Serine/threonine-protein Kinase 17B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.23 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.27 |
Binding ≤ 10μM
|
|
TBA1A-2-E |
Tubulin Alpha-3 Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM
|
|
ULK3-1-E |
Serine/threonine-protein Kinase ULK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.26 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.32 |
Functional ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
599 |
0.28 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
9800 |
0.23 |
Functional ≤ 10μM
|
|
Z80186-9-O |
K562 (Erythroleukemia Cells) (cluster #9 Of 11), Other |
Other |
9360 |
0.23 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1000 |
0.27 |
Functional ≤ 10μM
|
|
Z80262-1-O |
NCI-H1975 (Bronchoalveolar Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8260 |
0.23 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7400 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7000 |
0.23 |
Functional ≤ 10μM
|
|
Z80961-1-O |
HN5 (Squamous Cell Carcinoma) (cluster #1 Of 1), Other |
Other |
10 |
0.36 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
80 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
430 |
0.29 |
Binding ≤ 1μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
590 |
0.28 |
Binding ≤ 1μM
|
|
EPHB4_HUMAN |
P54760
|
Ephrin Type-B Receptor 4, Human |
1000 |
0.27 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
0.4 |
0.42 |
Binding ≤ 1μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
360 |
0.29 |
Binding ≤ 1μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
290 |
0.30 |
Binding ≤ 1μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
240 |
0.30 |
Binding ≤ 1μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
470 |
0.29 |
Binding ≤ 1μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
920 |
0.27 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
13 |
0.36 |
Binding ≤ 1μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
530 |
0.28 |
Binding ≤ 1μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
410 |
0.29 |
Binding ≤ 1μM
|
|
HCK_HUMAN |
P08631
|
Tyrosine-protein Kinase HCK, Human |
110 |
0.31 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
630 |
0.28 |
Binding ≤ 1μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
990 |
0.27 |
Binding ≤ 1μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
1000 |
0.27 |
Binding ≤ 1μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
1100 |
0.27 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
1400 |
0.26 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
2300 |
0.25 |
Binding ≤ 10μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
2000 |
0.26 |
Binding ≤ 10μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
1300 |
0.27 |
Binding ≤ 10μM
|
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
5700 |
0.24 |
Binding ≤ 10μM
|
|
EPHA1_HUMAN |
P21709
|
Ephrin Type-A Receptor 1, Human |
4000 |
0.24 |
Binding ≤ 10μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
1400 |
0.26 |
Binding ≤ 10μM
|
|
EPHA8_HUMAN |
P29322
|
Ephrin Type-A Receptor 8, Human |
1800 |
0.26 |
Binding ≤ 10μM
|
|
EPHB1_HUMAN |
P54762
|
Ephrin Type-B Receptor 1, Human |
7300 |
0.23 |
Binding ≤ 10μM
|
|
EPHB4_HUMAN |
P54760
|
Ephrin Type-B Receptor 4, Human |
1000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
0.4 |
0.42 |
Binding ≤ 10μM
|
|
IRAK3_HUMAN |
Q9Y616
|
Interleukin-1 Receptor-associated Kinase 3, Human |
1500 |
0.26 |
Binding ≤ 10μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
1200 |
0.27 |
Binding ≤ 10μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
290 |
0.30 |
Binding ≤ 10μM
|
|
MK04_HUMAN |
P31152
|
Mitogen-activated Protein Kinase 4, Human |
3100 |
0.25 |
Binding ≤ 10μM
|
|
MK06_HUMAN |
Q16659
|
Mitogen-activated Protein Kinase 6, Human |
1600 |
0.26 |
Binding ≤ 10μM
|
|
MYLK2_HUMAN |
Q9H1R3
|
Myosin Light Chain Kinase 2, Human |
1900 |
0.26 |
Binding ≤ 10μM
|
|
DMPK_HUMAN |
Q09013
|
Myotonin-protein Kinase, Human |
6900 |
0.23 |
Binding ≤ 10μM
|
|
PHKG1_HUMAN |
Q16816
|
Phosphorylase Kinase Gamma Subunit 1, Human |
3700 |
0.25 |
Binding ≤ 10μM
|
|
PHKG2_HUMAN |
P15735
|
Phosphorylase Kinase Gamma Subunit 2, Human |
6400 |
0.23 |
Binding ≤ 10μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
1100 |
0.27 |
Binding ≤ 10μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
470 |
0.29 |
Binding ≤ 10μM
|
|
ST17A_HUMAN |
Q9UEE5
|
Serine/threonine-protein Kinase 17A, Human |
2000 |
0.26 |
Binding ≤ 10μM
|
|
ST17B_HUMAN |
O94768
|
Serine/threonine-protein Kinase 17B, Human |
3800 |
0.24 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
1100 |
0.27 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
13 |
0.36 |
Binding ≤ 10μM
|
|
PIM3_HUMAN |
Q86V86
|
Serine/threonine-protein Kinase PIM3, Human |
5800 |
0.24 |
Binding ≤ 10μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
530 |
0.28 |
Binding ≤ 10μM
|
|
SIK2_HUMAN |
Q9H0K1
|
Serine/threonine-protein Kinase SIK2, Human |
2100 |
0.26 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
4300 |
0.24 |
Binding ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
410 |
0.29 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
1200 |
0.27 |
Binding ≤ 10μM
|
|
BLK_HUMAN |
P51451
|
Tyrosine-protein Kinase BLK, Human |
1200 |
0.27 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
2000 |
0.26 |
Binding ≤ 10μM
|
|
HCK_HUMAN |
P08631
|
Tyrosine-protein Kinase HCK, Human |
110 |
0.31 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
1100 |
0.27 |
Binding ≤ 10μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
990 |
0.27 |
Binding ≤ 10μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
1000 |
0.27 |
Binding ≤ 10μM
|
|
RET_HUMAN |
P07949
|
Tyrosine-protein Kinase Receptor RET, Human |
1700 |
0.26 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
1100 |
0.27 |
Binding ≤ 10μM
|
|
TXK_HUMAN |
P42681
|
Tyrosine-protein Kinase TXK, Human |
6000 |
0.24 |
Binding ≤ 10μM
|
|
ULK3_MOUSE |
Q3U3Q1
|
ULK3 Kinase, Mouse |
1400 |
0.26 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
1900 |
0.26 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
7000 |
0.23 |
Functional ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
14.4 |
0.35 |
Functional ≤ 10μM
|
|
Z80961 |
Z80961
|
HN5 (Squamous Cell Carcinoma) |
10 |
0.36 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
9800 |
0.23 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
9360 |
0.23 |
Functional ≤ 10μM
|
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
54 |
0.33 |
Functional ≤ 10μM
|
|
Z80188 |
Z80188
|
KB 3-1 (Cervical Epithelial Carcinoma Cells) |
380 |
0.29 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
1000 |
0.27 |
Functional ≤ 10μM
|
|
Z80262 |
Z80262
|
NCI-H1975 (Bronchoalveolar Carcinoma Cells) |
8260 |
0.23 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
7400 |
0.23 |
Functional ≤ 10μM
|
|
ERBB2_HUMAN |
P04626
|
Receptor Protein-tyrosine Kinase ErbB-2, Human |
1604 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
8.75 |
-11.24 |
1 |
7 |
0 |
69 |
446.91 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.93 |
9.92 |
-78.3 |
3 |
7 |
2 |
75 |
448.926 |
7 |
↓
|
|
|
|
Analogs
-
8214397
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
3.87 |
-1.64 |
0 |
0 |
0 |
0 |
197.381 |
1 |
↓
|
|
|
|
Analogs
-
34478674
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
3.08 |
-3.47 |
1 |
1 |
0 |
20 |
122.167 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.49 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
574 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-4-E |
GABA Receptor Alpha-1 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.58 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5 |
0.58 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
12 |
0.55 |
Binding ≤ 1μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
531 |
0.44 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA1_BOVIN |
P08219
|
GABA Receptor Alpha-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA2_BOVIN |
P10063
|
GABA Receptor Alpha-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
15 |
0.55 |
Binding ≤ 1μM
|
|
GBRA3_BOVIN |
P10064
|
GABA Receptor Alpha-3 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
13.9 |
0.55 |
Binding ≤ 1μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA4_BOVIN |
P20237
|
GABA Receptor Alpha-4 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
23 |
0.53 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
2.1 |
0.61 |
Binding ≤ 1μM
|
|
GBRB1_BOVIN |
P08220
|
GABA Receptor Beta-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRG2_BOVIN |
P22300
|
GABA Receptor Gamma-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10.5 |
0.56 |
Binding ≤ 1μM
|
|
TSPO_RAT |
P16257
|
Peripheral-type Benzodiazepine Receptor, Rat |
213 |
0.47 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL_RAT |
P02692
|
Fatty Acid-binding Protein, Liver, Rat |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA1_BOVIN |
P08219
|
GABA Receptor Alpha-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_BOVIN |
P10063
|
GABA Receptor Alpha-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_MOUSE |
P26048
|
GABA Receptor Alpha-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
15 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA3_BOVIN |
P10064
|
GABA Receptor Alpha-3 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
13.9 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_MOUSE |
P26049
|
GABA Receptor Alpha-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA4_MOUSE |
Q9D6F4
|
GABA Receptor Alpha-4 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA4_BOVIN |
P20237
|
GABA Receptor Alpha-4 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
23 |
0.53 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRA5_MOUSE |
Q8BHJ7
|
GABA Receptor Alpha-5 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA6_MOUSE |
P16305
|
GABA Receptor Alpha-6 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
2.1 |
0.61 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRB1_BOVIN |
P08220
|
GABA Receptor Beta-1 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB1_MOUSE |
P50571
|
GABA Receptor Beta-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRB3_MOUSE |
P63080
|
GABA Receptor Beta-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRD_MOUSE |
P22933
|
GABA Receptor Delta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
A2AMW3_MOUSE |
A2AMW3
|
GABA Receptor Epsilon Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1_MOUSE |
Q9R0Y8
|
GABA Receptor Gamma-1 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_MOUSE |
P22723
|
GABA Receptor Gamma-2 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRG2_BOVIN |
P22300
|
GABA Receptor Gamma-2 Subunit, Bovin |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG3_RAT |
P28473
|
GABA Receptor Gamma-3 Subunit, Rat |
11 |
0.56 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRG3_MOUSE |
P27681
|
GABA Receptor Gamma-3 Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
GBRP_MOUSE |
Q8QZW7
|
GABA Receptor Pi Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRT_MOUSE |
Q9JLF1
|
GABA Receptor Theta Subunit, Mouse |
12 |
0.55 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
4.9 |
0.58 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
10.5 |
0.56 |
Binding ≤ 10μM
|
|
TSPO_RAT |
P16257
|
Peripheral-type Benzodiazepine Receptor, Rat |
213 |
0.47 |
Binding ≤ 10μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
1.8 |
0.61 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
5 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
6.91 |
-8.26 |
0 |
3 |
0 |
33 |
284.746 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.74 |
7.78 |
-30.37 |
1 |
3 |
1 |
34 |
285.754 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
-1.86 |
-4.06 |
1 |
1 |
0 |
20 |
136.976 |
1 |
↓
|
|