|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
0.24 |
-67.99 |
5 |
11 |
-1 |
174 |
526.566 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
0.10 |
1.91 |
-47.63 |
6 |
11 |
0 |
175 |
527.574 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.29 |
-2.14 |
-141.04 |
4 |
11 |
-2 |
180 |
525.558 |
4 |
↓
|
|
|
|
Analogs
-
11592545
-
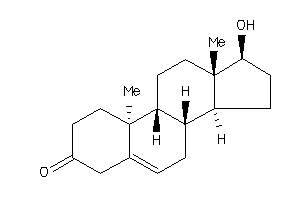
-
11592546
-
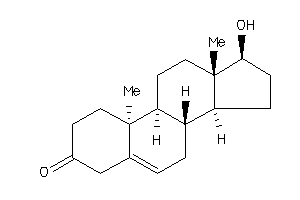
-
103270
-
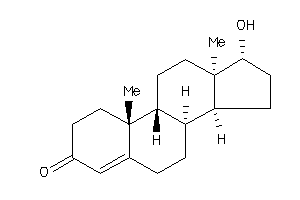
-
490793
-
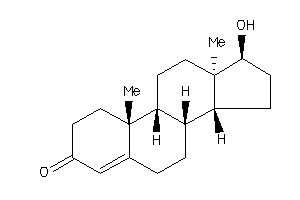
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 1μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 1μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 1μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
1.4 |
0.59 |
Binding ≤ 10μM
|
|
ANDR_HUMAN |
P10275
|
Androgen Receptor, Human |
2.7 |
0.57 |
Binding ≤ 10μM
|
|
ERG2_YEAST |
P32352
|
C-8 Sterol Isomerase, Yeast |
7760 |
0.34 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.41 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
1200 |
0.39 |
Binding ≤ 10μM
|
|
S5A1_RAT |
P24008
|
Steroid 5-alpha-reductase 1, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
S5A2_RAT |
P31214
|
Steroid 5-alpha-reductase 2, Rat |
1900 |
0.38 |
Binding ≤ 10μM
|
|
SHBG_HUMAN |
P04278
|
Testis-specific Androgen-binding Protein, Human |
14 |
0.52 |
Binding ≤ 10μM
|
|
ANDR_RAT |
P15207
|
Androgen Receptor, Rat |
3.2 |
0.57 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
10000 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.07 |
-7.05 |
1 |
2 |
0 |
37 |
288.431 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.99 |
5.16 |
-4.08 |
2 |
2 |
0 |
40 |
288.431 |
0 |
↓
|
|
|
|
Analogs
-
38735350
-
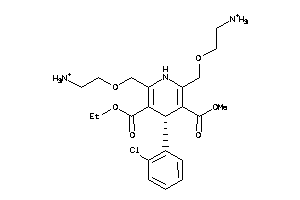
-
38735353
-
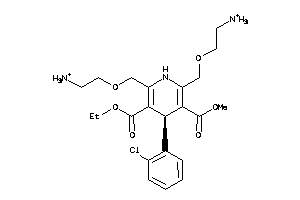
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2344 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7943 |
0.26 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5370 |
0.26 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
5350 |
0.26 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
2930 |
0.28 |
Functional ≤ 10μM
|
|
Z50458-3-O |
Leishmania Braziliensis (cluster #3 Of 4), Other |
Other |
3130 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-3-O |
Leishmania Major (cluster #3 Of 4), Other |
Other |
3350 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8070 |
0.25 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
7.61 |
-58.02 |
4 |
7 |
1 |
102 |
409.89 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.83 |
6.31 |
-39.45 |
2 |
7 |
-1 |
106 |
407.874 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
7.16 |
-13.89 |
3 |
7 |
0 |
100 |
408.882 |
10 |
↓
|
|
|
|
Analogs
-
38735350
-

-
38735353
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2344 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7943 |
0.26 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5370 |
0.26 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
5350 |
0.26 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
2930 |
0.28 |
Functional ≤ 10μM
|
|
Z50458-3-O |
Leishmania Braziliensis (cluster #3 Of 4), Other |
Other |
3130 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-3-O |
Leishmania Major (cluster #3 Of 4), Other |
Other |
3350 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8070 |
0.25 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
7.61 |
-60.61 |
4 |
7 |
1 |
102 |
409.89 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
7.17 |
-11.79 |
3 |
7 |
0 |
100 |
408.882 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.83 |
6.31 |
-39.43 |
2 |
7 |
-1 |
106 |
407.874 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH1-1-E |
Voltage-gated Potassium Channel Subunit Kv10.1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC9-2-E |
Sulfonylurea Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
|
IRK11-2-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
4 |
0.69 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
27 |
0.62 |
Binding ≤ 1μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
230 |
0.55 |
Binding ≤ 1μM
|
|
IRK11_HUMAN |
Q14654
|
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11, Human |
27 |
0.62 |
Binding ≤ 10μM
|
|
ABCC9_HUMAN |
O60706
|
Sulfonylurea Receptor 2, Human |
230 |
0.55 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
250 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
7.64 |
-8.48 |
2 |
5 |
0 |
73 |
231.303 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.88 |
9.38 |
-6.25 |
2 |
5 |
0 |
73 |
231.303 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.88 |
9.4 |
-8.38 |
2 |
5 |
0 |
73 |
231.303 |
5 |
↓
|
|
|
|
Analogs
-
33776615
-
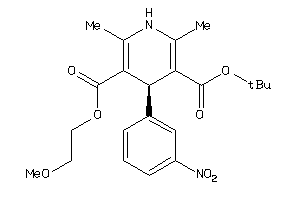
-
33776616
-
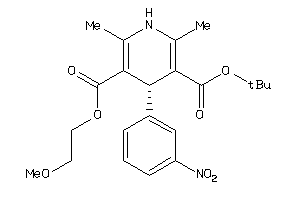
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8470 |
0.24 |
Binding ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.28 |
Functional ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.24 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
15 |
0.37 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 1), Other |
Other |
690 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
11.26 |
-18.9 |
1 |
9 |
0 |
120 |
418.446 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
80 |
0.50 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.46 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
128 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
0.68 |
0.64 |
Binding ≤ 1μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
123 |
0.48 |
Binding ≤ 1μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
28 |
0.53 |
Binding ≤ 1μM
|
|
ACES_MOUSE |
P21836
|
Acetylcholinesterase, Mouse |
0.68 |
0.64 |
Binding ≤ 1μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
21.3 |
0.54 |
Binding ≤ 1μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
1000 |
0.42 |
Binding ≤ 1μM
|
|
CHLE_MOUSE |
Q03311
|
Butyrylcholinesterase, Mouse |
280 |
0.46 |
Binding ≤ 1μM
|
|
CHLE_HUMAN |
P06276
|
Butyrylcholinesterase, Human |
16 |
0.55 |
Binding ≤ 1μM
|
|
Q9JKC1_RAT |
Q9JKC1
|
Butyrylcholinesterase, Rat |
23 |
0.53 |
Binding ≤ 1μM
|
|
CHLE_HORSE |
P81908
|
Cholinesterase, Horse |
857 |
0.42 |
Binding ≤ 1μM
|
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
0.68 |
0.64 |
Binding ≤ 10μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
123 |
0.48 |
Binding ≤ 10μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
28 |
0.53 |
Binding ≤ 10μM
|
|
ACES_MOUSE |
P21836
|
Acetylcholinesterase, Mouse |
0.68 |
0.64 |
Binding ≤ 10μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
21.3 |
0.54 |
Binding ≤ 10μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
1000 |
0.42 |
Binding ≤ 10μM
|
|
CHLE_MOUSE |
Q03311
|
Butyrylcholinesterase, Mouse |
280 |
0.46 |
Binding ≤ 10μM
|
|
CHLE_HUMAN |
P06276
|
Butyrylcholinesterase, Human |
16 |
0.55 |
Binding ≤ 10μM
|
|
Q9JKC1_RAT |
Q9JKC1
|
Butyrylcholinesterase, Rat |
23 |
0.53 |
Binding ≤ 10μM
|
|
CHLE_HORSE |
P81908
|
Cholinesterase, Horse |
1340 |
0.41 |
Binding ≤ 10μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
128 |
0.48 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
-0.07 |
-7.08 |
1 |
5 |
0 |
48 |
275.352 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
2.61 |
-39.01 |
2 |
5 |
1 |
50 |
276.36 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
4.54 |
-76.78 |
3 |
5 |
2 |
51 |
277.368 |
2 |
↓
|
|
|
|
Analogs
-
34319816
-
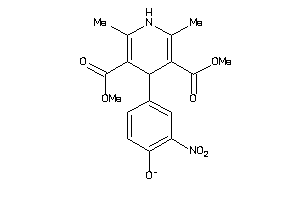
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8300 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.29 |
Functional ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.27 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80387-3-O |
PC-12 (Adrenal Phaeochromacytoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
8300 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10000 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80387 |
Z80387
|
PC-12 (Adrenal Phaeochromacytoma Cells) |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.27 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
28 |
0.41 |
Functional ≤ 10μM
|
|
TRPA1_MOUSE |
Q8BLA8
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Mouse |
3800 |
0.29 |
Functional ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
9.32 |
-35.55 |
0 |
8 |
-1 |
117 |
359.358 |
6 |
↓
|
|
|
|
Analogs
-
34319816
-

-
597260
-
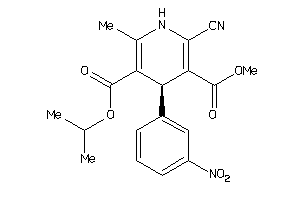
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8300 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.29 |
Functional ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.27 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80387-3-O |
PC-12 (Adrenal Phaeochromacytoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
0.15 |
0.53 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
8300 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10000 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
0.15 |
0.53 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80387 |
Z80387
|
PC-12 (Adrenal Phaeochromacytoma Cells) |
10 |
0.43 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.27 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
28 |
0.41 |
Functional ≤ 10μM
|
|
TRPA1_MOUSE |
Q8BLA8
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Mouse |
3800 |
0.29 |
Functional ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F_HUMAN |
O60840
|
Voltage-gated L-type Calcium Channel Alpha-1F Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S_HUMAN |
Q13698
|
Voltage-gated L-type Calcium Channel Alpha-1S Subunit, Human |
1 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
10.4 |
-17.04 |
1 |
8 |
0 |
110 |
360.366 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.74 |
-46.91 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
4.54 |
-14.78 |
2 |
7 |
0 |
74 |
427.545 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
6.89 |
-46.27 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.81 |
-48.71 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
6.92 |
-48.11 |
3 |
7 |
1 |
75 |
428.553 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.92 |
4.61 |
-16.16 |
2 |
7 |
0 |
74 |
427.545 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8100 |
0.31 |
ADME/T ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
2 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
9.54 |
-32.05 |
1 |
3 |
1 |
31 |
326.782 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.07 |
9.07 |
-10.63 |
0 |
3 |
0 |
30 |
325.774 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.07 |
10 |
-33.25 |
1 |
3 |
1 |
32 |
326.782 |
1 |
↓
|
|
|
|
Analogs
-
4676424
-
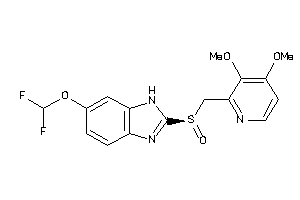
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
2.48 |
-24.9 |
1 |
7 |
0 |
86 |
383.376 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
1.95 |
2.49 |
-24.17 |
1 |
7 |
0 |
86 |
383.376 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.95 |
2.02 |
-54.17 |
0 |
7 |
-1 |
85 |
382.368 |
7 |
↓
|
|
|
|
Analogs
-
4099200
-
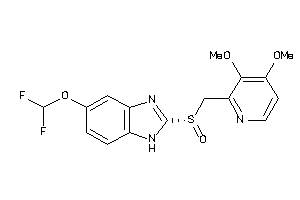
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
2.48 |
-16.82 |
1 |
7 |
0 |
86 |
383.376 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.95 |
2.01 |
-55.16 |
0 |
7 |
-1 |
85 |
382.368 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.95 |
2.01 |
-55.75 |
0 |
7 |
-1 |
85 |
382.368 |
7 |
↓
|
|
|
|
Analogs
-
21984495
-
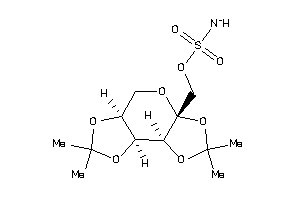
-
22061272
-
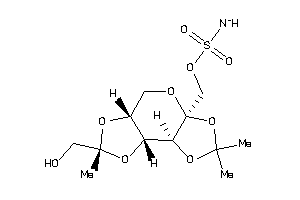
-
22061274
-
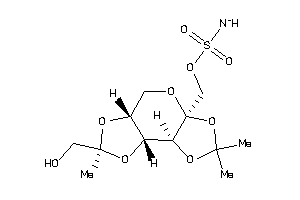
-
23586802
-
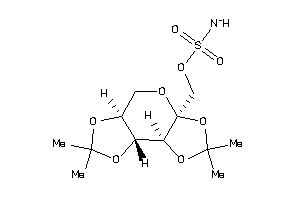
-
23586806
-
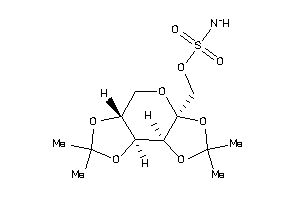
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-2-A |
Carbonic Anhydrase (cluster #2 Of 2), Archaea |
Archaea |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CYNT-3-B |
Carbonic Anhydrase (cluster #3 Of 3), Bacterial |
Bacteria |
474 |
0.40 |
Binding ≤ 10μM
|
|
P96878-2-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #2 Of 2), Bacterial |
Bacteria |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284-2-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #2 Of 2), Bacterial |
Bacteria |
612 |
0.40 |
Binding ≤ 10μM
|
|
B5SU02-4-E |
Alpha Carbonic Anhydrase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
C0IX24-3-E |
Carbonic Anhydrase (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
367 |
0.41 |
Binding ≤ 10μM
|
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-3-E |
Carbonic Anhydrase XIII (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14-5-E |
Carbonic Anhydrase XIV (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1460 |
0.37 |
Binding ≤ 10μM
|
|
CAH15-5-E |
Carbonic Anhydrase 15 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH3-4-E |
Carbonic Anhydrase III (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-7-E |
Carbonic Anhydrase IV (cluster #7 Of 16), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH5A-3-E |
Carbonic Anhydrase VA (cluster #3 Of 10), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5B-3-E |
Carbonic Anhydrase VB (cluster #3 Of 9), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-5-E |
Carbonic Anhydrase VII (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
58 |
0.46 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2-2-E |
Carbonic Anhydrase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4-2-E |
Carbonic Anhydrase IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Functional ≤ 10μM
|
|
CAN-2-F |
Carbonic Anhydrase (cluster #2 Of 3), Fungal |
Fungi |
110 |
0.44 |
Binding ≤ 10μM
|
|
Q3I4V7-4-F |
Carbonic Anhydrase 2 (cluster #4 Of 4), Fungal |
Fungi |
370 |
0.41 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
1110 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
29.1 |
0.48 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
110 |
0.44 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
367 |
0.41 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
120 |
0.44 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
10 |
0.51 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
474 |
0.40 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
172 |
0.43 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.45 |
Binding ≤ 1μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
370 |
0.41 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
15 |
0.50 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
58 |
0.46 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
25.4 |
0.48 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
63 |
0.46 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
25.4 |
0.48 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
63 |
0.46 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
45 |
0.47 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.87 |
0.58 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3.5 |
0.54 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
47 |
0.47 |
Binding ≤ 1μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
47 |
0.47 |
Binding ≤ 1μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
610 |
0.40 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
29.1 |
0.48 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
10 |
0.51 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
110 |
0.44 |
Binding ≤ 10μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
474 |
0.40 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
1020 |
0.38 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
367 |
0.41 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
172 |
0.43 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.45 |
Binding ≤ 10μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
370 |
0.41 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
15 |
0.50 |
Binding ≤ 10μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4900 |
0.34 |
Binding ≤ 10μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
1590 |
0.37 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
63 |
0.46 |
Binding ≤ 10μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
25.4 |
0.48 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
25.4 |
0.48 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
63 |
0.46 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
1360 |
0.37 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.87 |
0.58 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3.5 |
0.54 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
1460 |
0.37 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
610 |
0.40 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
54 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
-1.52 |
-44.79 |
1 |
9 |
-1 |
113 |
338.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.16 |
-2 |
-12.26 |
2 |
9 |
0 |
116 |
339.366 |
3 |
↓
|
|
|
|
Analogs
-
35572125
-
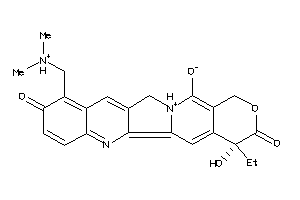
-
35643838
-
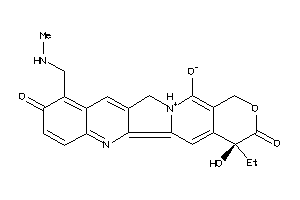
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HIF1A-1-E |
Hypoxia-inducible Factor 1-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.33 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.33 |
Binding ≤ 10μM
|
|
Z100519-1-O |
KETR3 (Renal Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
13 |
0.36 |
Functional ≤ 10μM
|
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
49 |
0.33 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
300 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3800 |
0.24 |
Functional ≤ 10μM
|
|
Z80105-1-O |
COR-L23 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.35 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
1200 |
0.27 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
63 |
0.33 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
74 |
0.32 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
52 |
0.33 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
880 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
1700 |
0.26 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
440 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
601 |
0.28 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.34 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5550 |
0.24 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
38 |
0.34 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
31 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
26 |
0.34 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
140 |
0.31 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
48 |
0.33 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
150 |
0.31 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
1500 |
0.26 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
500 |
0.28 |
Functional ≤ 10μM
|
|
Z80596-1-O |
WISH (Amniotic Epithelial Cells) (cluster #1 Of 1), Other |
Other |
1300 |
0.27 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
160 |
0.31 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
87 |
0.32 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
78 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
104 |
0.32 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
21 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1100 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
56 |
0.33 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.33 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
7100 |
0.23 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8700 |
0.23 |
Functional ≤ 10μM
|
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
339 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
70 |
0.32 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
9 |
0.36 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
79 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1600 |
0.26 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.27 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.38 |
Functional ≤ 10μM
|
|
Z81342-1-O |
Tumour Cell Lines (cluster #1 Of 1), Other |
Other |
40 |
0.33 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
3100 |
0.25 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.39 |
-40.99 |
3 |
8 |
0 |
108 |
422.461 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
4.23 |
-51.04 |
2 |
8 |
0 |
107 |
421.453 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
2.6 |
-55.53 |
2 |
8 |
0 |
105 |
421.453 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
Z81047-1-O |
Human T-cell Line (cluster #1 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
0.22 |
-59.92 |
3 |
5 |
0 |
85 |
272.37 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.82 |
-1.07 |
-41.57 |
2 |
5 |
-1 |
80 |
271.362 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
0.05 |
-49.18 |
4 |
5 |
1 |
83 |
273.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.40 |
Binding ≤ 10μM
|
|
Z81047-1-O |
Human T-cell Line (cluster #1 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
0.82 |
-0.91 |
-41.79 |
2 |
5 |
-1 |
80 |
271.362 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
8.77 |
-7.56 |
1 |
3 |
0 |
38 |
359.665 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
5.94 |
8.77 |
-7.17 |
1 |
3 |
0 |
38 |
359.665 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Lo
Low (pH 4.5-6)
|
4.12 |
9.72 |
-41.19 |
3 |
9 |
0 |
134 |
446.511 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.12 |
9.62 |
-36.92 |
3 |
9 |
0 |
134 |
446.511 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
1438
-
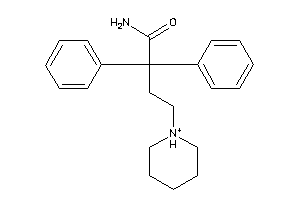
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.47 |
10.33 |
-39.34 |
2 |
3 |
1 |
43 |
337.487 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2360 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.29 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
106 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1585 |
0.29 |
Binding ≤ 10μM
|
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
1 |
0.45 |
Binding ≤ 10μM
|
|
Z50597-5-O |
Rattus Norvegicus (cluster #5 Of 5), Other |
Other |
3000 |
0.28 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.86 |
0.45 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
0.1 |
0.50 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.254 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
0.27 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.17 |
0.49 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
0.19 |
0.49 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.21 |
0.48 |
Binding ≤ 1μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.83 |
0.45 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
106 |
0.35 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 1μM
|
|
MT3_HUMAN |
P25713
|
Metallothionein-3, Human |
7.8 |
0.41 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 1μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
0.55 |
0.46 |
Binding ≤ 1μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
0.33 |
0.47 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.86 |
0.45 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
0.1 |
0.50 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
0.27 |
0.48 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
0.17 |
0.49 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.254 |
0.48 |
Binding ≤ 10μM
|
|
ADA1B_MESAU |
P18841
|
Alpha-1b Adrenergic Receptor, Mesau |
0.83 |
0.45 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
0.19 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.21 |
0.48 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
ADA2A_PIG |
P18871
|
Alpha-2a Adrenergic Receptor, Pig |
4100 |
0.27 |
Binding ≤ 10μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
106 |
0.35 |
Binding ≤ 10μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
108 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
13 |
0.39 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.4 |
0.47 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
8300 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1584.89319 |
0.29 |
Binding ≤ 10μM
|
|
MT3_HUMAN |
P25713
|
Metallothionein-3, Human |
7.8 |
0.41 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.2 |
0.48 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
3000 |
0.28 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
2360 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B_HUMAN |
P28222
|
Serotonin 1b (5-HT1b) Receptor, Human |
0.55 |
0.46 |
Binding ≤ 10μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
0.33 |
0.47 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1500 |
0.29 |
Binding ≤ 10μM
|
|
S22A1_HUMAN |
O15245
|
Solute Carrier Family 22 Member 1, Human |
9900 |
0.25 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.25 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
0.13 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
7.64 |
-36.73 |
3 |
9 |
1 |
108 |
384.416 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
7.26 |
-15.54 |
2 |
9 |
0 |
107 |
383.408 |
4 |
↓
|
|
|
|
Analogs
-
598034
-
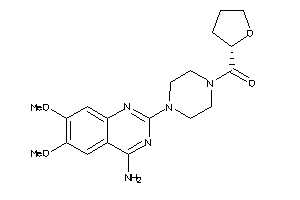
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.38 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.31 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.36 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.82 |
0.45 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2 |
0.43 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
26 |
0.38 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.69 |
0.46 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
1.9 |
0.44 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.85 |
0.45 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
1.01 |
0.45 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
550 |
0.31 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
7.7 |
0.41 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
26 |
0.38 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
170 |
0.34 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.82 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2 |
0.43 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
26 |
0.38 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.69 |
0.46 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
1.9 |
0.44 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.85 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
1.01 |
0.45 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
1500 |
0.29 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
7.7 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
26 |
0.38 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
170 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.8 |
-42.42 |
3 |
9 |
1 |
104 |
388.448 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.31 |
7.48 |
-18.61 |
2 |
9 |
0 |
103 |
387.44 |
4 |
↓
|
|
|
|
Analogs
-
1271441
-
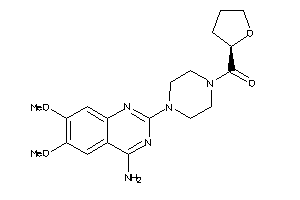
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.38 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.31 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.36 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.82 |
0.45 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2 |
0.43 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
26 |
0.38 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.69 |
0.46 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
1.9 |
0.44 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.85 |
0.45 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
1.01 |
0.45 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
550 |
0.31 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
7.7 |
0.41 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
26 |
0.38 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
170 |
0.34 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.82 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
2 |
0.43 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
26 |
0.38 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
0.69 |
0.46 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
1.9 |
0.44 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
0.85 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
1.01 |
0.45 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
1500 |
0.29 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
7.7 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
26 |
0.38 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
170 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.78 |
-39.71 |
3 |
9 |
1 |
104 |
388.448 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.31 |
7.48 |
-19.52 |
2 |
9 |
0 |
103 |
387.44 |
4 |
↓
|
|
|
|
Analogs
-
1802
-
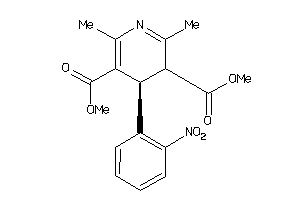
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2890 |
0.31 |
Binding ≤ 10μM
|
|
AA2AR-4-E |
Adenosine A2a Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8290 |
0.28 |
Binding ≤ 10μM
|
|
CAC1A-1-E |
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4300 |
0.30 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Functional ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
ADME/T ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 1), Other |
Other |
1 |
0.50 |
Binding ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
20 |
0.43 |
Functional ≤ 10μM
|
|
Z102372-1-O |
Aorta (cluster #1 Of 1), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM |
|
Z102380-2-O |
Ileum (cluster #2 Of 3), Other |
Other |
15 |
0.44 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
7 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM
|
|
Z80051-1-O |
C6-BU-1 (cluster #1 Of 1), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
1 |
0.50 |
Binding ≤ 1μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1C_RABIT |
P15381
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rabit |
1 |
0.50 |
Binding ≤ 1μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.4 |
0.53 |
Binding ≤ 1μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
2.5 |
0.48 |
Binding ≤ 1μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1B_RAT |
Q02294
|
Voltage-gated N-type Calcium Channel Alpha-1B Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1A_RAT |
P54282
|
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1E_RAT |
Q07652
|
Voltage-gated R-type Calcium Channel Alpha-1E Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1G_RAT |
O54898
|
Voltage-gated T-type Calcium Channel Alpha-1G Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1H_RAT |
Q9EQ60
|
Voltage-gated T-type Calcium Channel Alpha-1H Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
CAC1I_RAT |
Q9Z0Y8
|
Voltage-gated T-type Calcium Channel Alpha-1I Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
10000 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
4300 |
0.30 |
Binding ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
1 |
0.50 |
Binding ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
0.4 |
0.53 |
Binding ≤ 10μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1C_RABIT |
P15381
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rabit |
1 |
0.50 |
Binding ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
2.5 |
0.48 |
Binding ≤ 10μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1B_RAT |
Q02294
|
Voltage-gated N-type Calcium Channel Alpha-1B Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1A_RAT |
P54282
|
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
KCNA5_HUMAN |
P22460
|
Voltage-gated Potassium Channel Subunit Kv1.5, Human |
6100 |
0.29 |
Binding ≤ 10μM
|
|
CAC1E_RAT |
Q07652
|
Voltage-gated R-type Calcium Channel Alpha-1E Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1G_RAT |
O54898
|
Voltage-gated T-type Calcium Channel Alpha-1G Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1H_RAT |
Q9EQ60
|
Voltage-gated T-type Calcium Channel Alpha-1H Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
CAC1I_RAT |
Q9Z0Y8
|
Voltage-gated T-type Calcium Channel Alpha-1I Subunit, Rat |
0.53 |
0.52 |
Binding ≤ 10μM
|
|
Z102372 |
Z102372
|
Aorta |
9 |
0.45 |
Functional ≤ 10μM |
|
Z102306 |
Z102306
|
Aorta |
20 |
0.43 |
Functional ≤ 10μM
|
|
Z80051 |
Z80051
|
C6-BU-1 |
1000 |
0.34 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
14 |
0.44 |
Functional ≤ 10μM
|
|
Z102380 |
Z102380
|
Ileum |
14.7 |
0.44 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
0.5 |
0.52 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.29 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1.16 |
0.50 |
Functional ≤ 10μM
|
|
Z50590 |
Z50590
|
Sus Scrofa |
3.6 |
0.47 |
Functional ≤ 10μM
|
|
TRPA1_MOUSE |
Q8BLA8
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Mouse |
400 |
0.36 |
Functional ≤ 10μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
2.53 |
0.48 |
Functional ≤ 10μM
|
|
CAC1C_RABIT |
P15381
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rabit |
2.9 |
0.48 |
Functional ≤ 10μM
|
|
CAC1C_HUMAN |
Q13936
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Human |
2.5 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D_HUMAN |
Q01668
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Human |
2.5 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D_RAT |
P27732
|
Voltage-gated L-type Calcium Channel Alpha-1D Subunit, Rat |
2.53 |
0.48 |
Functional ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
10000 |
0.28 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
8.22 |
-42.09 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.32 |
7.84 |
-40.28 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.55 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.55 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.55 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
74 |
0.50 |
Binding ≤ 10μM
|
|
ADCY2-2-E |
Adenylate Cyclase Type II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY3-2-E |
Adenylate Cyclase Type III (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ADCY4-2-E |
Adenylate Cyclase Type IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY5-2-E |
Adenylate Cyclase Type V (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ADCY6-2-E |
Adenylate Cyclase Type VI (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.52 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
768 |
0.43 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
Q8CFM9-1-E |
Adenylate Cyclase Type VII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
3981 |
0.38 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.66 |
0.64 |
Binding ≤ 1μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ADCY5_HUMAN |
O95622
|
Adenylate Cyclase Type V, Human |
1.3 |
0.62 |
Binding ≤ 1μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
0.47 |
0.65 |
Binding ≤ 1μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
74 |
0.50 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
74 |
0.50 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
0.12 |
0.69 |
Binding ≤ 1μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
DRD1_MOUSE |
Q61616
|
Dopamine D1 Receptor, Mouse |
0.4 |
0.66 |
Binding ≤ 1μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
0.5 |
0.65 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
0.17 |
0.68 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
1000 |
0.42 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.42 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
768 |
0.43 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
0.48 |
0.65 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
10.8 |
0.56 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
13 |
0.55 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
12.51 |
0.55 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
9.2 |
0.56 |
Binding ≤ 1μM
|
|
SC6A4_MOUSE |
Q60857
|
Serotonin Transporter, Mouse |
9.2 |
0.56 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
0.66 |
0.64 |
Binding ≤ 10μM
|
|
ADCY2_RAT |
P26769
|
Adenylate Cyclase Type II, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ADCY3_RAT |
P21932
|
Adenylate Cyclase Type III, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ADCY4_RAT |
P26770
|
Adenylate Cyclase Type IV, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ADCY5_RAT |
Q04400
|
Adenylate Cyclase Type V, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ADCY5_HUMAN |
O95622
|
Adenylate Cyclase Type V, Human |
1.3 |
0.62 |
Binding ≤ 10μM
|
|
ADCY6_RAT |
Q03343
|
Adenylate Cyclase Type VI, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
Q8CFM9_RAT |
Q8CFM9
|
Adenylate Cyclase Type VII, Rat |
0.47 |
0.65 |
Binding ≤ 10μM
|
|
ADCY8_RAT |
P40146
|
Adenylate Cyclase Type VIII, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
74 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
74 |
0.50 |
Binding ≤ 10μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
0.5 |
0.65 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
0.17 |
0.68 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
0.12 |
0.69 |
Binding ≤ 10μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
DRD1_MOUSE |
Q61616
|
Dopamine D1 Receptor, Mouse |
0.4 |
0.66 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_MOUSE |
P61168
|
Dopamine D2 Receptor, Mouse |
1210 |
0.41 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
1000 |
0.42 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
768 |
0.43 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
0.48 |
0.65 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
10.8 |
0.56 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
13 |
0.55 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
12.51 |
0.55 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
9.2 |
0.56 |
Binding ≤ 10μM
|
|
SC6A4_MOUSE |
Q60857
|
Serotonin Transporter, Mouse |
9.2 |
0.56 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.35 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
0.28 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
8.28 |
-39.17 |
2 |
2 |
1 |
25 |
288.798 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.89 |
9.05 |
-61.19 |
1 |
2 |
0 |
27 |
287.79 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.89 |
8.63 |
-45.42 |
0 |
2 |
-1 |
26 |
286.782 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
732 |
0.33 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
284 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
8.91 |
-53.21 |
2 |
3 |
1 |
38 |
345.466 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.24 |
9.29 |
-91.65 |
3 |
3 |
2 |
39 |
346.474 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
6.12 |
-47.64 |
5 |
5 |
1 |
87 |
416.582 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.94 |
4.71 |
-9.45 |
4 |
5 |
0 |
82 |
415.574 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
6.19 |
-46.77 |
5 |
5 |
1 |
87 |
416.582 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.94 |
4.83 |
-9.23 |
4 |
5 |
0 |
82 |
415.574 |
16 |
↓
|
|
|
|
Analogs
-
33748262
-
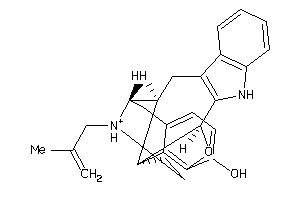
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
6.13 |
-47.35 |
4 |
5 |
1 |
70 |
415.513 |
2 |
↓
|
|
|
|
Analogs
-
5820938
-
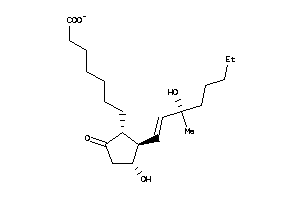
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R1-2-E |
Prostanoid EP1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
PE2R2-2-E |
Prostanoid EP2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.41 |
Binding ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
PE2R4-2-E |
Prostanoid EP4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
PI2R-2-E |
Prostanoid IP Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.38 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
70 |
0.40 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R1_MOUSE |
P35375
|
Prostanoid EP1 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 1μM
|
|
PE2R2_MOUSE |
Q62053
|
Prostanoid EP2 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 1μM
|
|
PE2R3_MOUSE |
P30557
|
Prostanoid EP3 Receptor, Mouse |
5 |
0.46 |
Binding ≤ 1μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
3.1 |
0.48 |
Binding ≤ 1μM
|
|
PI2R_HUMAN |
P43119
|
Prostanoid IP Receptor, Human |
150 |
0.38 |
Binding ≤ 1μM
|
|
PE2R1_MOUSE |
P35375
|
Prostanoid EP1 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 10μM
|
|
PE2R2_MOUSE |
Q62053
|
Prostanoid EP2 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 10μM
|
|
PE2R3_MOUSE |
P30557
|
Prostanoid EP3 Receptor, Mouse |
5 |
0.46 |
Binding ≤ 10μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
3.1 |
0.48 |
Binding ≤ 10μM
|
|
PI2R_HUMAN |
P43119
|
Prostanoid IP Receptor, Human |
1400 |
0.33 |
Binding ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
70 |
0.40 |
Functional ≤ 10μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
2.5 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
7.89 |
-49.67 |
2 |
5 |
-1 |
98 |
353.479 |
13 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.36 |
5.91 |
-10.15 |
3 |
5 |
0 |
95 |
354.487 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
19 |
0.45 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
339 |
0.38 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
123 |
0.40 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1318 |
0.34 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1318 |
0.34 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1318 |
0.34 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1318 |
0.34 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1318 |
0.34 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.30 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
19 |
0.45 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
46.7735141 |
0.43 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
150 |
0.40 |
Binding ≤ 1μM
|
|
5HT1D_PIG |
P79400
|
Serotonin 1d (5-HT1d) Receptor, Pig |
123.026877 |
0.40 |
Binding ≤ 1μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
35 |
0.43 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
19 |
0.45 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
46.7735141 |
0.43 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
150 |
0.40 |
Binding ≤ 10μM
|
|
5HT1D_PIG |
P79400
|
Serotonin 1d (5-HT1d) Receptor, Pig |
123.026877 |
0.40 |
Binding ≤ 10μM
|
|
5HT1D_HUMAN |
P28221
|
Serotonin 1d (5-HT1d) Receptor, Human |
35 |
0.43 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
6800 |
0.30 |
Binding ≤ 10μM
|
|
5HT3A_HUMAN |
P46098
|
Serotonin 3a (5-HT3a) Receptor, Human |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3B_HUMAN |
O95264
|
Serotonin 3b (5-HT3b) Receptor, Human |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3C_HUMAN |
Q8WXA8
|
Serotonin 3c (5-HT3c) Receptor, Human |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3D_HUMAN |
Q70Z44
|
Serotonin 3d (5-HT3d) Receptor, Human |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
5HT3E_HUMAN |
A5X5Y0
|
Serotonin 3e (5-HT3e) Receptor, Human |
1318.25674 |
0.34 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.6 |
-44.27 |
1 |
4 |
1 |
25 |
335.353 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.62 |
8.17 |
-8.23 |
0 |
4 |
0 |
24 |
334.345 |
2 |
↓
|
|
|
|
Analogs
-
8830624
-
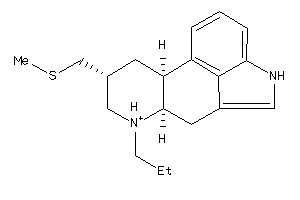
-
33884202
-
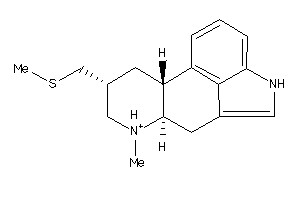
-
1909
-
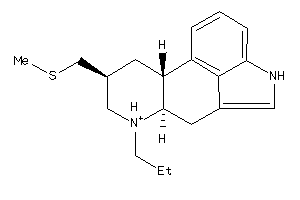
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
70 |
0.46 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
50 |
0.46 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
70 |
0.46 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.34 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
0.1 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
11.27 |
-39.66 |
2 |
2 |
1 |
20 |
315.506 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.97 |
9.08 |
-5.67 |
1 |
2 |
0 |
19 |
314.498 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
5.33 |
-52.38 |
0 |
6 |
-1 |
96 |
369.422 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.31 |
7.06 |
-16.88 |
1 |
6 |
0 |
89 |
370.43 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
10.41 |
-42.03 |
1 |
2 |
1 |
22 |
264.364 |
5 |
↓
|
|
|
|
Analogs
-
38413756
-
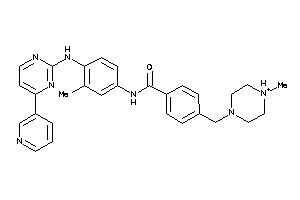
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199-1-E |
BCR/ABL P210 Fusion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.23 |
Binding ≤ 10μM
|
|
ABCG2-2-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4898 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.24 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
468 |
0.24 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
30 |
0.28 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
76 |
0.27 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.20 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.20 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.23 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.28 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.30 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.19 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.21 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.27 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.25 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.27 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.19 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.22 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.20 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Functional ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.19 |
Functional ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
9100 |
0.19 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
6 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
10000 |
0.19 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199_HUMAN |
A1Z199
|
BCR/ABL P210 Fusion Protein, Human |
204 |
0.25 |
Binding ≤ 1μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
1.1 |
0.34 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.27 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
31.9 |
0.28 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
30.2 |
0.28 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
528 |
0.24 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
75.7 |
0.27 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
392 |
0.24 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
109 |
0.26 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
980 |
0.23 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
468 |
0.24 |
Binding ≤ 1μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
141 |
0.26 |
Binding ≤ 1μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
0.7 |
0.35 |
Binding ≤ 1μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
19 |
0.29 |
Binding ≤ 1μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
160 |
0.26 |
Binding ≤ 1μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
14 |
0.30 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
1000 |
0.23 |
Binding ≤ 1μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
100 |
0.26 |
Binding ≤ 1μM
|
|
ABL1_MOUSE |
P00520
|
Tyrosine-protein Kinase ABL, Mouse |
10.8 |
0.30 |
Binding ≤ 1μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
430 |
0.24 |
Binding ≤ 1μM
|
|
ABL2_HUMAN |
P42684
|
Tyrosine-protein Kinase ABL2, Human |
10 |
0.30 |
Binding ≤ 1μM
|
|
BLK_HUMAN |
P51451
|
Tyrosine-protein Kinase BLK, Human |
520 |
0.24 |
Binding ≤ 1μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
160 |
0.26 |
Binding ≤ 1μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
220 |
0.25 |
Binding ≤ 1μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
3380 |
0.21 |
Binding ≤ 10μM
|
|
A1Z199_HUMAN |
A1Z199
|
BCR/ABL P210 Fusion Protein, Human |
204 |
0.25 |
Binding ≤ 10μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
1.1 |
0.34 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
3200 |
0.21 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
3100 |
0.21 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
31.9 |
0.28 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
30.2 |
0.28 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
75.7 |
0.27 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
468 |
0.24 |
Binding ≤ 10μM
|
|
CDK19_HUMAN |
Q9BWU1
|
Cell Division Cycle 2-like Protein Kinase 6, Human |
5500 |
0.20 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
141 |
0.26 |
Binding ≤ 10μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
2100 |
0.21 |
Binding ≤ 10μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
4500 |
0.20 |
Binding ≤ 10μM
|
|
EPHA8_HUMAN |
P29322
|
Ephrin Type-A Receptor 8, Human |
1400 |
0.22 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
7600 |
0.19 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
0.7 |
0.35 |
Binding ≤ 10μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
1274 |
0.22 |
Binding ≤ 10μM
|
|
MELK_HUMAN |
Q14680
|
Maternal Embryonic Leucine Zipper Kinase, Human |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MLTK_HUMAN |
Q9NYL2
|
Mixed Lineage Kinase 7, Human |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
160 |
0.26 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
14 |
0.30 |
Binding ≤ 10μM
|
|
ST17A_HUMAN |
Q9UEE5
|
Serine/threonine-protein Kinase 17A, Human |
2800 |
0.21 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
3300 |
0.21 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
1000 |
0.23 |
Binding ≤ 10μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
7800 |
0.19 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
1700 |
0.22 |
Binding ≤ 10μM
|
|
TNI3K_HUMAN |
Q59H18
|
Serine/threonine-protein Kinase TNNI3K, Human |
4300 |
0.20 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
100 |
0.26 |
Binding ≤ 10μM
|
|
ABL1_MOUSE |
P00520
|
Tyrosine-protein Kinase ABL, Mouse |
10.8 |
0.30 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
430 |
0.24 |
Binding ≤ 10μM
|
|
ABL2_HUMAN |
P42684
|
Tyrosine-protein Kinase ABL2, Human |
10 |
0.30 |
Binding ≤ 10μM
|
|
BLK_HUMAN |
P51451
|
Tyrosine-protein Kinase BLK, Human |
520 |
0.24 |
Binding ≤ 10μM
|
|
FGR_HUMAN |
P09769
|
Tyrosine-protein Kinase FGR, Human |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
1500 |
0.22 |
Binding ≤ 10μM
|
|
FYN_HUMAN |
P06241
|
Tyrosine-protein Kinase FYN, Human |
3100 |
0.21 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
160 |
0.26 |
Binding ≤ 10μM
|
|
LYN_HUMAN |
P07948
|
Tyrosine-protein Kinase Lyn, Human |
220 |
0.25 |
Binding ≤ 10μM
|
|
KSYK_HUMAN |
P43405
|
Tyrosine-protein Kinase SYK, Human |
5000 |
0.20 |
Binding ≤ 10μM
|
|
Z80036 |
Z80036
|
BaF3 (IL-3-dependent Pro-B-cells) |
400 |
0.24 |
Functional ≤ 10μM
|
|
BCR_HUMAN |
P11274
|
Breakpoint Cluster Region Protein, Human |
90 |
0.27 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
10000 |
0.19 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
90 |
0.27 |
Functional ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
96 |
0.27 |
Functional ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
219 |
0.25 |
Functional ≤ 10μM
|
|
Z50466 |
Z50466
|
Trypanosoma Cruzi |
9000 |
0.19 |
Functional ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
90 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
13.15 |
-49.4 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
13.03 |
-52.36 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
10.78 |
-14.7 |
2 |
8 |
0 |
86 |
493.615 |
7 |
↓
|
|
|
|
Analogs
-
1536786
-
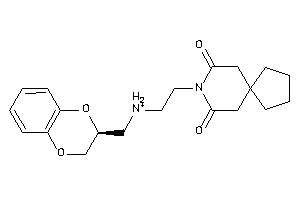
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
7.83 |
-43.72 |
2 |
6 |
1 |
72 |
359.446 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.45 |
6.47 |
-11.81 |
1 |
6 |
0 |
68 |
358.438 |
5 |
↓
|
|
|
|
Analogs
-
1999423
-
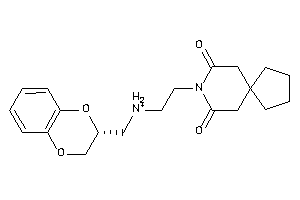
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
7.85 |
-43.46 |
2 |
6 |
1 |
72 |
359.446 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.45 |
6.48 |
-12.63 |
1 |
6 |
0 |
68 |
358.438 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
-1.89 |
-54.12 |
4 |
3 |
1 |
60 |
243.33 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.11 |
-3.29 |
-50.81 |
4 |
3 |
1 |
60 |
243.33 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
HRH4-2-E |
Histamine H4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
0.338844156 |
0.66 |
Binding ≤ 1μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
0.18 |
0.68 |
Binding ≤ 1μM
|
|
HRH3_MOUSE |
P58406
|
Histamine H3 Receptor, Mouse |
0.6 |
0.65 |
Binding ≤ 1μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
10 |
0.56 |
Binding ≤ 1μM
|
|
HRH3_MOUSE |
P58406
|
Histamine H3 Receptor, Mouse |
0.6 |
0.65 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
0.338844156 |
0.66 |
Binding ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
0.18 |
0.68 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
10 |
0.56 |
Binding ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
0.58 |
0.65 |
Functional ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
0.18 |
0.68 |
Functional ≤ 10μM
|
|
HRH3_MOUSE |
P58406
|
Histamine H3 Receptor, Mouse |
1 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
8.47 |
-84.45 |
5 |
4 |
2 |
70 |
310.854 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.67 |
8.11 |
-8.6 |
3 |
4 |
0 |
65 |
308.838 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.67 |
8.13 |
-38.57 |
4 |
4 |
1 |
66 |
309.846 |
8 |
↓
|
|
|
|
Analogs
-
531272
-
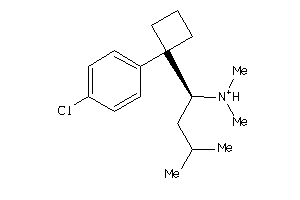
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.53 |
-34.64 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
Analogs
-
4759
-
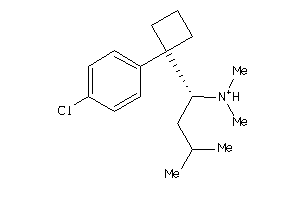
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.74 |
-33.26 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
10 |
-42.36 |
2 |
2 |
1 |
29 |
311.836 |
0 |
↓
|
|