|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
5.66 |
-32.76 |
2 |
8 |
1 |
87 |
361.422 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.74 |
5.16 |
-92.9 |
7 |
8 |
2 |
122 |
438.507 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.84 |
5.67 |
-39.11 |
2 |
8 |
1 |
87 |
361.422 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
7.12 |
-10.35 |
2 |
6 |
0 |
80 |
311.37 |
5 |
↓
|
|
|
|
Analogs
-
13122806
-
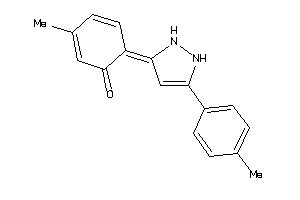
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
6.13 |
-8.58 |
2 |
3 |
0 |
49 |
250.301 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.82 |
6.92 |
-49.24 |
1 |
3 |
-1 |
52 |
249.293 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.82 |
6.77 |
-47.27 |
1 |
3 |
-1 |
52 |
249.293 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
-2.48 |
-15.9 |
3 |
5 |
0 |
86 |
286.283 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.6 |
-11.91 |
2 |
7 |
0 |
82 |
391.422 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.93 |
8.31 |
-50.21 |
1 |
7 |
-1 |
84 |
390.414 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.93 |
7.81 |
-39.47 |
3 |
7 |
1 |
83 |
392.43 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.63 |
-11.57 |
2 |
7 |
0 |
82 |
391.422 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.93 |
8.35 |
-50.44 |
1 |
7 |
-1 |
84 |
390.414 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.93 |
7.83 |
-40.47 |
3 |
7 |
1 |
83 |
392.43 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AVR2B-1-E |
Activin Receptor Type-2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
92 |
0.34 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.27 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.29 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.33 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AVR2B_HUMAN |
Q13705
|
Activin Receptor Type-2B, Human |
92 |
0.34 |
Binding ≤ 1μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
780 |
0.29 |
Binding ≤ 1μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
3.7 |
0.41 |
Binding ≤ 1μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
17 |
0.38 |
Binding ≤ 1μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
160 |
0.33 |
Binding ≤ 1μM
|
|
AVR2B_HUMAN |
Q13705
|
Activin Receptor Type-2B, Human |
92 |
0.34 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
2600 |
0.27 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
780 |
0.29 |
Binding ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
3.7 |
0.41 |
Binding ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
17 |
0.38 |
Binding ≤ 10μM
|
|
PI4KB_HUMAN |
Q9UBF8
|
PI4-kinase Beta Subunit, Human |
3300 |
0.26 |
Binding ≤ 10μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
160 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
5.98 |
-25.03 |
4 |
6 |
0 |
102 |
404.323 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
130 |
0.36 |
Binding ≤ 1μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
290 |
0.34 |
Binding ≤ 1μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
120 |
0.36 |
Binding ≤ 1μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
100 |
0.36 |
Binding ≤ 1μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
420 |
0.33 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
340 |
0.34 |
Binding ≤ 1μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
1000 |
0.31 |
Binding ≤ 1μM
|
|
MK14_MOUSE |
P47811
|
MAP Kinase P38 Alpha, Mouse |
136 |
0.36 |
Binding ≤ 1μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
Q5SC61_TOXGO |
Q5SC61
|
Mitogen-activated Protein Kinase 1, Toxgo |
100 |
0.36 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
320 |
0.34 |
Binding ≤ 1μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
250 |
0.34 |
Binding ≤ 1μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
25 |
0.39 |
Binding ≤ 1μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
860 |
0.31 |
Binding ≤ 1μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
530 |
0.33 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
135 |
0.36 |
Binding ≤ 1μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
400 |
0.33 |
Binding ≤ 1μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
330 |
0.34 |
Binding ≤ 1μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
16 |
0.40 |
Binding ≤ 1μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
820 |
0.32 |
Binding ≤ 1μM
|
|
ACV1B_HUMAN |
P36896
|
Activin Receptor Type-1B, Human |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
130 |
0.36 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
290 |
0.34 |
Binding ≤ 10μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
120 |
0.36 |
Binding ≤ 10μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
100 |
0.36 |
Binding ≤ 10μM
|
|
KC1AL_HUMAN |
Q8N752
|
Casein Kinase I Isoform Alpha-like, Human |
1700 |
0.30 |
Binding ≤ 10μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
420 |
0.33 |
Binding ≤ 10μM
|
|
PGH1_HUMAN |
P23219
|
Cyclooxygenase-1, Human |
2000 |
0.30 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
MP2K2_HUMAN |
P36507
|
Dual Specificity Mitogen-activated Protein Kinase Kinase 2, Human |
9600 |
0.26 |
Binding ≤ 10μM
|
|
TTK_HUMAN |
P33981
|
Dual Specificity Protein Kinase TTK, Human |
2500 |
0.29 |
Binding ≤ 10μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2_HUMAN |
P29323
|
Ephrin Type-B Receptor 2, Human |
6800 |
0.27 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
1300 |
0.31 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
1000 |
0.31 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
1700 |
0.30 |
Binding ≤ 10μM
|
|
MK14_MOUSE |
P47811
|
MAP Kinase P38 Alpha, Mouse |
136 |
0.36 |
Binding ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
Q5SC61_TOXGO |
Q5SC61
|
Mitogen-activated Protein Kinase 1, Toxgo |
100 |
0.36 |
Binding ≤ 10μM
|
|
M4K4_HUMAN |
O95819
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4, Human |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MLTK_HUMAN |
Q9NYL2
|
Mixed Lineage Kinase 7, Human |
4400 |
0.28 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
250 |
0.34 |
Binding ≤ 10μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
25 |
0.39 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
3700 |
0.28 |
Binding ≤ 10μM
|
|
ST32B_HUMAN |
Q9NY57
|
Serine/threonine-protein Kinase 32B, Human |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
1300 |
0.31 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
530 |
0.33 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
135 |
0.36 |
Binding ≤ 10μM
|
|
MRCKB_HUMAN |
Q9Y5S2
|
Serine/threonine-protein Kinase MRCK Beta, Human |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
400 |
0.33 |
Binding ≤ 10μM
|
|
MRCKA_HUMAN |
Q5VT25
|
Serine/threonine-protein Kinase MRCK-A, Human |
6200 |
0.27 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
2000 |
0.30 |
Binding ≤ 10μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
16 |
0.40 |
Binding ≤ 10μM
|
|
TNI3K_HUMAN |
Q59H18
|
Serine/threonine-protein Kinase TNNI3K, Human |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TGFR1_HUMAN |
P36897
|
TGF-beta Receptor Type I, Human |
6000 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2_HUMAN |
P37173
|
TGF-beta Receptor Type II, Human |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
1500 |
0.30 |
Binding ≤ 10μM
|
|
PTK6_HUMAN |
Q13882
|
Tyrosine-protein Kinase BRK, Human |
3900 |
0.28 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
1800 |
0.30 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
2800 |
0.29 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
TXK_HUMAN |
P42681
|
Tyrosine-protein Kinase TXK, Human |
4500 |
0.28 |
Binding ≤ 10μM
|
|
ADCK4_HUMAN |
Q96D53
|
Uncharacterized AarF Domain-containing Protein Kinase 4, Human |
3100 |
0.29 |
Binding ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
80 |
0.37 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
2000 |
0.30 |
Functional ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
160 |
0.35 |
Functional ≤ 10μM
|
|
Z80523 |
Z80523
|
SW1353 |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548 |
Z80548
|
THP-1 (Acute Monocytic Leukemia Cells) |
16 |
0.40 |
Functional ≤ 10μM
|
|
TNFA_HUMAN |
P01375
|
TNF-alpha, Human |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z102116 |
Z102116
|
Toxoplasma Gondii RH |
8500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-21.46 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.75 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.24 |
-46.38 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
130 |
0.36 |
Binding ≤ 1μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
290 |
0.34 |
Binding ≤ 1μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
120 |
0.36 |
Binding ≤ 1μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
100 |
0.36 |
Binding ≤ 1μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
420 |
0.33 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
340 |
0.34 |
Binding ≤ 1μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
1000 |
0.31 |
Binding ≤ 1μM
|
|
MK14_MOUSE |
P47811
|
MAP Kinase P38 Alpha, Mouse |
136 |
0.36 |
Binding ≤ 1μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
10 |
0.41 |
Binding ≤ 1μM
|
|
Q5SC61_TOXGO |
Q5SC61
|
Mitogen-activated Protein Kinase 1, Toxgo |
100 |
0.36 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
320 |
0.34 |
Binding ≤ 1μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
250 |
0.34 |
Binding ≤ 1μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
25 |
0.39 |
Binding ≤ 1μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
860 |
0.31 |
Binding ≤ 1μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
530 |
0.33 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
135 |
0.36 |
Binding ≤ 1μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
400 |
0.33 |
Binding ≤ 1μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
330 |
0.34 |
Binding ≤ 1μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
16 |
0.40 |
Binding ≤ 1μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
820 |
0.32 |
Binding ≤ 1μM
|
|
ACV1B_HUMAN |
P36896
|
Activin Receptor Type-1B, Human |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
130 |
0.36 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
290 |
0.34 |
Binding ≤ 10μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
120 |
0.36 |
Binding ≤ 10μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
100 |
0.36 |
Binding ≤ 10μM
|
|
KC1AL_HUMAN |
Q8N752
|
Casein Kinase I Isoform Alpha-like, Human |
1700 |
0.30 |
Binding ≤ 10μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
420 |
0.33 |
Binding ≤ 10μM
|
|
PGH1_HUMAN |
P23219
|
Cyclooxygenase-1, Human |
2000 |
0.30 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
MP2K2_HUMAN |
P36507
|
Dual Specificity Mitogen-activated Protein Kinase Kinase 2, Human |
9600 |
0.26 |
Binding ≤ 10μM
|
|
TTK_HUMAN |
P33981
|
Dual Specificity Protein Kinase TTK, Human |
2500 |
0.29 |
Binding ≤ 10μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2_HUMAN |
P29323
|
Ephrin Type-B Receptor 2, Human |
6800 |
0.27 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
1300 |
0.31 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
1000 |
0.31 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
1700 |
0.30 |
Binding ≤ 10μM
|
|
MK14_MOUSE |
P47811
|
MAP Kinase P38 Alpha, Mouse |
136 |
0.36 |
Binding ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
10 |
0.41 |
Binding ≤ 10μM
|
|
Q5SC61_TOXGO |
Q5SC61
|
Mitogen-activated Protein Kinase 1, Toxgo |
100 |
0.36 |
Binding ≤ 10μM
|
|
M4K4_HUMAN |
O95819
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4, Human |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MLTK_HUMAN |
Q9NYL2
|
Mixed Lineage Kinase 7, Human |
4400 |
0.28 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
250 |
0.34 |
Binding ≤ 10μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
25 |
0.39 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
3700 |
0.28 |
Binding ≤ 10μM
|
|
ST32B_HUMAN |
Q9NY57
|
Serine/threonine-protein Kinase 32B, Human |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
1300 |
0.31 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
530 |
0.33 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
135 |
0.36 |
Binding ≤ 10μM
|
|
MRCKB_HUMAN |
Q9Y5S2
|
Serine/threonine-protein Kinase MRCK Beta, Human |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
400 |
0.33 |
Binding ≤ 10μM
|
|
MRCKA_HUMAN |
Q5VT25
|
Serine/threonine-protein Kinase MRCK-A, Human |
6200 |
0.27 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
2000 |
0.30 |
Binding ≤ 10μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
16 |
0.40 |
Binding ≤ 10μM
|
|
TNI3K_HUMAN |
Q59H18
|
Serine/threonine-protein Kinase TNNI3K, Human |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TGFR1_HUMAN |
P36897
|
TGF-beta Receptor Type I, Human |
6000 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2_HUMAN |
P37173
|
TGF-beta Receptor Type II, Human |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
1500 |
0.30 |
Binding ≤ 10μM
|
|
PTK6_HUMAN |
Q13882
|
Tyrosine-protein Kinase BRK, Human |
3900 |
0.28 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
1800 |
0.30 |
Binding ≤ 10μM
|
|
LCK_HUMAN |
P06239
|
Tyrosine-protein Kinase LCK, Human |
2800 |
0.29 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
TXK_HUMAN |
P42681
|
Tyrosine-protein Kinase TXK, Human |
4500 |
0.28 |
Binding ≤ 10μM
|
|
ADCK4_HUMAN |
Q96D53
|
Uncharacterized AarF Domain-containing Protein Kinase 4, Human |
3100 |
0.29 |
Binding ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
80 |
0.37 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
2000 |
0.30 |
Functional ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
1000 |
0.31 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
160 |
0.35 |
Functional ≤ 10μM
|
|
Z80523 |
Z80523
|
SW1353 |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548 |
Z80548
|
THP-1 (Acute Monocytic Leukemia Cells) |
16 |
0.40 |
Functional ≤ 10μM
|
|
TNFA_HUMAN |
P01375
|
TNF-alpha, Human |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z102116 |
Z102116
|
Toxoplasma Gondii RH |
8500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.81 |
-16.64 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-16.98 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.22 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
AURKA-2-E |
Serine/threonine-protein Kinase Aurora-A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
760 |
0.31 |
Binding ≤ 10μM |
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
390 |
0.32 |
Binding ≤ 10μM
|
|
AURKC-2-E |
Serine/threonine-protein Kinase Aurora-C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.36 |
Binding ≤ 10μM
|
|
BMPR2-1-E |
Bone Morphogenetic Protein Receptor Type-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
CD11A-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
|
CD11B-1-E |
PITSLRE Serine/threonine-protein Kinase CDC2L1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9800 |
0.25 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.36 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.28 |
Binding ≤ 10μM
|
|
CDK8-1-E |
Cell Division Protein Kinase 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.25 |
Binding ≤ 10μM
|
|
CLK4-2-E |
Dual Specificity Protein Kinase CLK4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
58 |
0.36 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6500 |
0.26 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.32 |
Binding ≤ 10μM
|
|
EPHB3-1-E |
Ephrin Type-B Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
INSRR-1-E |
Insulin Receptor-related Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3900 |
0.27 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6900 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
81 |
0.35 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.26 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
LIMK2-1-E |
LIM Domain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.27 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.31 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
830 |
0.30 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.27 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
910 |
0.30 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.30 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.33 |
Binding ≤ 10μM
|
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.27 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.33 |
Binding ≤ 10μM
|
|
MUSK-1-E |
Muscle, Skeletal Receptor Tyrosine Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
|
NTRK3-1-E |
NT-3 Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.33 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.36 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8500 |
0.25 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
RIPK1-1-E |
Receptor-interacting Serine/threonine-protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.33 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.32 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.29 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.27 |
Binding ≤ 10μM
|
|
STK4-2-E |
Serine/threonine-protein Kinase MST1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.35 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.32 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.34 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
240 |
0.33 |
Binding ≤ 1μM
|
|
CDK19_HUMAN |
Q9BWU1
|
Cell Division Cycle 2-like Protein Kinase 6, Human |
74 |
0.36 |
Binding ≤ 1μM
|
|
CDK8_HUMAN |
P49336
|
Cell Division Protein Kinase 8, Human |
95 |
0.35 |
Binding ≤ 1μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
330 |
0.32 |
Binding ≤ 1μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
58 |
0.36 |
Binding ≤ 1μM
|
|
LTK_HUMAN |
P29376
|
Leukocyte Tyrosine Kinase Receptor, Human |
550 |
0.31 |
Binding ≤ 1μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
3 |
0.43 |
Binding ≤ 1μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
270 |
0.33 |
Binding ≤ 1μM
|
|
M4K1_HUMAN |
Q92918
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1, Human |
830 |
0.30 |
Binding ≤ 1μM
|
|
M4K5_HUMAN |
Q9Y4K4
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5, Human |
910 |
0.30 |
Binding ≤ 1μM
|
|
MUSK_HUMAN |
O15146
|
Muscle, Skeletal Receptor Tyrosine Protein Kinase, Human |
10 |
0.40 |
Binding ≤ 1μM
|
|
NTRK3_HUMAN |
Q16288
|
NT-3 Growth Factor Receptor, Human |
210 |
0.33 |
Binding ≤ 1μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
4.2 |
0.42 |
Binding ≤ 1μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
1.9 |
0.44 |
Binding ≤ 1μM
|
|
MERTK_HUMAN |
Q12866
|
Proto-oncogene Tyrosine-protein Kinase MER, Human |
870 |
0.30 |
Binding ≤ 1μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
460 |
0.32 |
Binding ≤ 1μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
450 |
0.32 |
Binding ≤ 1μM
|
|
ST32B_HUMAN |
Q9NY57
|
Serine/threonine-protein Kinase 32B, Human |
270 |
0.33 |
Binding ≤ 1μM
|
|
AURKA_HUMAN |
O14965
|
Serine/threonine-protein Kinase Aurora-A, Human |
760 |
0.31 |
Binding ≤ 1μM |
|
AURKB_HUMAN |
Q96GD4
|
Serine/threonine-protein Kinase Aurora-B, Human |
390 |
0.32 |
Binding ≤ 1μM
|
|
AURKC_HUMAN |
Q9UQB9
|
Serine/threonine-protein Kinase Aurora-C, Human |
71 |
0.36 |
Binding ≤ 1μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
1.7 |
0.44 |
Binding ≤ 1μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
400 |
0.32 |
Binding ≤ 1μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
0.63 |
0.46 |
Binding ≤ 1μM
|
|
RET_HUMAN |
P07949
|
Tyrosine-protein Kinase Receptor RET, Human |
100 |
0.35 |
Binding ≤ 1μM
|
|
TIE1_HUMAN |
P35590
|
Tyrosine-protein Kinase Receptor Tie-1, Human |
110 |
0.35 |
Binding ≤ 1μM
|
|
UFO_HUMAN |
P30530
|
Tyrosine-protein Kinase Receptor UFO, Human |
340 |
0.32 |
Binding ≤ 1μM
|
|
TIE2_HUMAN |
Q02763
|
Tyrosine-protein Kinase TIE-2, Human |
170 |
0.34 |
Binding ≤ 1μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
3 |
0.43 |
Binding ≤ 1μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
28 |
0.38 |
Binding ≤ 1μM
|
|
VGFR3_HUMAN |
P35916
|
Vascular Endothelial Growth Factor Receptor 3, Human |
16 |
0.39 |
Binding ≤ 1μM
|
|
ALK_HUMAN |
Q9UM73
|
ALK Tyrosine Kinase Receptor, Human |
1600 |
0.29 |
Binding ≤ 10μM
|
|
BMPR2_HUMAN |
Q13873
|
Bone Morphogenetic Protein Receptor Type-2, Human |
7800 |
0.26 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
240 |
0.33 |
Binding ≤ 10μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
6900 |
0.26 |
Binding ≤ 10μM
|
|
CDK19_HUMAN |
Q9BWU1
|
Cell Division Cycle 2-like Protein Kinase 6, Human |
74 |
0.36 |
Binding ≤ 10μM
|
|
CDK8_HUMAN |
P49336
|
Cell Division Protein Kinase 8, Human |
95 |
0.35 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
9800 |
0.25 |
Binding ≤ 10μM
|
|
CDK7_HUMAN |
P50613
|
Cyclin-dependent Kinase 7, Human |
2900 |
0.28 |
Binding ≤ 10μM
|
|
CDK9_HUMAN |
P50750
|
Cyclin-dependent Kinase 9, Human |
3000 |
0.28 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
3800 |
0.27 |
Binding ≤ 10μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CLK1_HUMAN |
P49759
|
Dual Specificty Protein Kinase CLK1, Human |
8900 |
0.25 |
Binding ≤ 10μM
|
|
EPHA3_HUMAN |
P29320
|
Ephrin Type-A Receptor 3, Human |
3500 |
0.27 |
Binding ≤ 10μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
330 |
0.32 |
Binding ≤ 10μM
|
|
EPHB3_HUMAN |
P54753
|
Ephrin Type-B Receptor 3, Human |
1400 |
0.29 |
Binding ≤ 10μM
|
|
EPHB4_HUMAN |
P54760
|
Ephrin Type-B Receptor 4, Human |
1400 |
0.29 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
6500 |
0.26 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
58 |
0.36 |
Binding ≤ 10μM
|
|
MET_HUMAN |
P08581
|
Hepatocyte Growth Factor Receptor, Human |
1300 |
0.29 |
Binding ≤ 10μM
|
|
INSRR_HUMAN |
P14616
|
Insulin Receptor-related Protein, Human |
3900 |
0.27 |
Binding ≤ 10μM
|
|
LTK_HUMAN |
P29376
|
Leukocyte Tyrosine Kinase Receptor, Human |
550 |
0.31 |
Binding ≤ 10μM
|
|
LIMK2_HUMAN |
P53671
|
LIM Domain Kinase 2, Human |
4700 |
0.27 |
Binding ≤ 10μM
|
|
CSF1R_HUMAN |
P07333
|
Macrophage Colony Stimulating Factor Receptor, Human |
3 |
0.43 |
Binding ≤ 10μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
270 |
0.33 |
Binding ≤ 10μM
|
|
MKNK1_HUMAN |
Q9BUB5
|
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1, Human |
4900 |
0.27 |
Binding ≤ 10μM
|
|
MK15_HUMAN |
Q8TD08
|
Mitogen-activated Protein Kinase 15, Human |
2800 |
0.28 |
Binding ≤ 10μM
|
|
M4K1_HUMAN |
Q92918
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1, Human |
830 |
0.30 |
Binding ≤ 10μM
|
|
M4K3_HUMAN |
Q8IVH8
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3, Human |
3200 |
0.27 |
Binding ≤ 10μM
|
|
M4K5_HUMAN |
Q9Y4K4
|
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5, Human |
910 |
0.30 |
Binding ≤ 10μM
|
|
MUSK_HUMAN |
O15146
|
Muscle, Skeletal Receptor Tyrosine Protein Kinase, Human |
10 |
0.40 |
Binding ≤ 10μM
|
|
NTRK1_HUMAN |
P04629
|
Nerve Growth Factor Receptor Trk-A, Human |
3100 |
0.28 |
Binding ≤ 10μM
|
|
NTRK2_HUMAN |
Q16620
|
Neurotrophic Tyrosine Kinase Receptor Type 2, Human |
1100 |
0.30 |
Binding ≤ 10μM
|
|
NTRK3_HUMAN |
Q16288
|
NT-3 Growth Factor Receptor, Human |
210 |
0.33 |
Binding ≤ 10μM
|
|
CD11B_HUMAN |
P21127
|
PITSLRE Serine/threonine-protein Kinase CDC2L1, Human |
1300 |
0.29 |
Binding ≤ 10μM
|
|
CD11A_HUMAN |
Q9UQ88
|
PITSLRE Serine/threonine-protein Kinase CDC2L2, Human |
1200 |
0.30 |
Binding ≤ 10μM
|
|
PGFRA_HUMAN |
P16234
|
Platelet-derived Growth Factor Receptor Alpha, Human |
4.2 |
0.42 |
Binding ≤ 10μM
|
|
PGFRB_HUMAN |
P09619
|
Platelet-derived Growth Factor Receptor Beta, Human |
1.9 |
0.44 |
Binding ≤ 10μM
|
|
MERTK_HUMAN |
Q12866
|
Proto-oncogene Tyrosine-protein Kinase MER, Human |
870 |
0.30 |
Binding ≤ 10μM
|
|
RIPK1_HUMAN |
Q13546
|
Receptor-interacting Serine/threonine-protein Kinase 1, Human |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KS6A4_HUMAN |
O75676
|
Ribosomal Protein S6 Kinase Alpha 4, Human |
7200 |
0.26 |
Binding ≤ 10μM
|
|
KS6A5_HUMAN |
O75582
|
Ribosomal Protein S6 Kinase Alpha 5, Human |
1200 |
0.30 |
Binding ≤ 10μM
|
|
STK10_HUMAN |
O94804
|
Serine/threonine-protein Kinase 10, Human |
460 |
0.32 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
450 |
0.32 |
Binding ≤ 10μM
|
|
ST32B_HUMAN |
Q9NY57
|
Serine/threonine-protein Kinase 32B, Human |
270 |
0.33 |
Binding ≤ 10μM
|
|
STK33_HUMAN |
Q9BYT3
|
Serine/threonine-protein Kinase 33, Human |
3800 |
0.27 |
Binding ≤ 10μM
|
|
AURKA_HUMAN |
O14965
|
Serine/threonine-protein Kinase Aurora-A, Human |
1600 |
0.29 |
Binding ≤ 10μM
|
|
AURKB_HUMAN |
Q96GD4
|
Serine/threonine-protein Kinase Aurora-B, Human |
390 |
0.32 |
Binding ≤ 10μM
|
|
AURKC_HUMAN |
Q9UQB9
|
Serine/threonine-protein Kinase Aurora-C, Human |
71 |
0.36 |
Binding ≤ 10μM
|
|
LATS2_HUMAN |
Q9NRM7
|
Serine/threonine-protein Kinase LATS2, Human |
1700 |
0.29 |
Binding ≤ 10μM
|
|
STK4_HUMAN |
Q13043
|
Serine/threonine-protein Kinase MST1, Human |
1400 |
0.29 |
Binding ≤ 10μM
|
|
STK3_HUMAN |
Q13188
|
Serine/threonine-protein Kinase MST2, Human |
1500 |
0.29 |
Binding ≤ 10μM
|
|
PLK3_HUMAN |
Q9H4B4
|
Serine/threonine-protein Kinase PLK3, Human |
8500 |
0.25 |
Binding ≤ 10μM
|
|
KIT_HUMAN |
P10721
|
Stem Cell Growth Factor Receptor, Human |
1.7 |
0.44 |
Binding ≤ 10μM
|
|
ABL1_HUMAN |
P00519
|
Tyrosine-protein Kinase ABL, Human |
1700 |
0.29 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
400 |
0.32 |
Binding ≤ 10μM
|
|
FLT3_HUMAN |
P36888
|
Tyrosine-protein Kinase Receptor FLT3, Human |
0.63 |
0.46 |
Binding ≤ 10μM
|
|
RET_HUMAN |
P07949
|
Tyrosine-protein Kinase Receptor RET, Human |
100 |
0.35 |
Binding ≤ 10μM
|
|
TIE1_HUMAN |
P35590
|
Tyrosine-protein Kinase Receptor Tie-1, Human |
110 |
0.35 |
Binding ≤ 10μM
|
|
UFO_HUMAN |
P30530
|
Tyrosine-protein Kinase Receptor UFO, Human |
340 |
0.32 |
Binding ≤ 10μM
|
|
TIE2_HUMAN |
Q02763
|
Tyrosine-protein Kinase TIE-2, Human |
170 |
0.34 |
Binding ≤ 10μM
|
|
VGFR1_HUMAN |
P17948
|
Vascular Endothelial Growth Factor Receptor 1, Human |
3 |
0.43 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
28 |
0.38 |
Binding ≤ 10μM
|
|
VGFR3_HUMAN |
P35916
|
Vascular Endothelial Growth Factor Receptor 3, Human |
16 |
0.39 |
Binding ≤ 10μM
|
|
VGFR2_HUMAN |
P35968
|
Vascular Endothelial Growth Factor Receptor 2, Human |
4 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
7.88 |
-12.09 |
5 |
6 |
0 |
96 |
375.407 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
9.9 |
-8.2 |
0 |
3 |
0 |
25 |
328.415 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.64 |
10.25 |
-30.61 |
1 |
3 |
1 |
26 |
329.423 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
9.87 |
-8.21 |
0 |
3 |
0 |
25 |
328.415 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.64 |
10.24 |
-30.63 |
1 |
3 |
1 |
26 |
329.423 |
0 |
↓
|
|
|
|
Analogs
-
538644
-
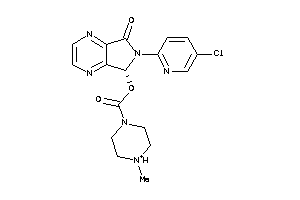
-
601241
-
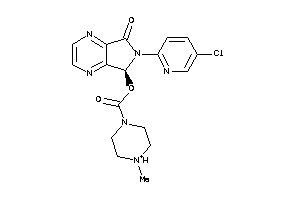
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
29 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.33 |
-15.82 |
0 |
9 |
0 |
92 |
388.815 |
3 |
↓
|
|
|
|
Analogs
-
538644
-

-
601241
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
114 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
162 |
0.35 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
102 |
0.36 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
29 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.34 |
-16.14 |
0 |
9 |
0 |
92 |
388.815 |
3 |
↓
|
|
|
|
Analogs
-
32131087
-
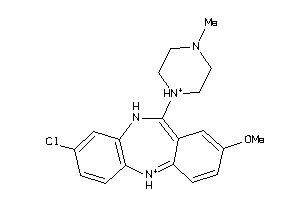
-
32131089
-
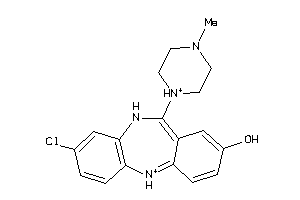
-
3995781
-
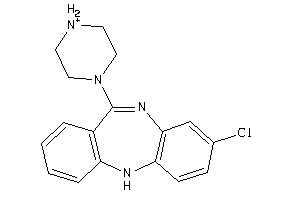
-
4292831
-
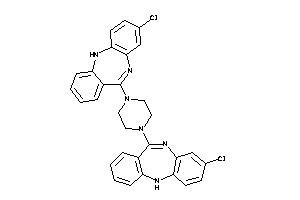
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
156 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.31 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.37 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4-1-E |
Histamine H4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.32 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.39 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
697 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.31 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
631 |
0.38 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
191 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
51 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.49 |
Binding ≤ 10μM
|
|
Z102275-1-O |
Hypothalamus (cluster #1 Of 1), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
156 |
0.41 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.54 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
114.815362 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
132 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
420 |
0.39 |
Binding ≤ 1μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
DRD2_MOUSE |
P61168
|
Dopamine D2 Receptor, Mouse |
290 |
0.40 |
Binding ≤ 1μM
|
|
DRD2_CHLAE |
P52702
|
Dopamine D2 Receptor, Chlae |
254 |
0.40 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
113 |
0.42 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
140 |
0.42 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
230 |
0.40 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
247 |
0.40 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
247 |
0.40 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
190.546072 |
0.41 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.1 |
0.55 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
14 |
0.48 |
Binding ≤ 1μM
|
|
HRH2_RAT |
P25102
|
Histamine H2 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.9 |
0.48 |
Binding ≤ 1μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
0.48 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
2.1 |
0.53 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.98 |
0.55 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
46 |
0.45 |
Binding ≤ 1μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
91 |
0.43 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
91 |
0.43 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
697 |
0.37 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
140 |
0.42 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
111 |
0.42 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT2A_MOUSE |
P35363
|
Serotonin 2a (5-HT2a) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
12 |
0.48 |
Binding ≤ 1μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
3 |
0.52 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2B_MOUSE |
Q02152
|
Serotonin 2b (5-HT2b) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
12 |
0.48 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
5HT2C_MOUSE |
P34968
|
Serotonin 2c (5-HT2c) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
53 |
0.44 |
Binding ≤ 1μM
|
|
5HT3A_MOUSE |
P23979
|
Serotonin 3a (5-HT3a) Receptor, Mouse |
32 |
0.46 |
Binding ≤ 1μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
53 |
0.44 |
Binding ≤ 1μM
|
|
5HT5A_HUMAN |
P47898
|
Serotonin 5a (5-HT5a) Receptor, Human |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
39.8107171 |
0.45 |
Binding ≤ 1μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
6.3 |
0.50 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
390 |
0.39 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
156 |
0.41 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.54 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
114.815362 |
0.42 |
Binding ≤ 10μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
420 |
0.39 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
132 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
DRD2_MOUSE |
P61168
|
Dopamine D2 Receptor, Mouse |
290 |
0.40 |
Binding ≤ 10μM
|
|
DRD2_CHLAE |
P52702
|
Dopamine D2 Receptor, Chlae |
254 |
0.40 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
113 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
140 |
0.42 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
230 |
0.40 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
247 |
0.40 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
247 |
0.40 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
190.546072 |
0.41 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.1 |
0.55 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
14 |
0.48 |
Binding ≤ 10μM
|
|
HRH2_RAT |
P25102
|
Histamine H2 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.9 |
0.48 |
Binding ≤ 10μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
0.48 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
2.1 |
0.53 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.98 |
0.55 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
46 |
0.45 |
Binding ≤ 10μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
697 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
140 |
0.42 |
Binding ≤ 10μM
|
|
5HT1A_MOUSE |
Q64264
|
Serotonin 1a (5-HT1a) Receptor, Mouse |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
1010 |
0.36 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT2A_MOUSE |
P35363
|
Serotonin 2a (5-HT2a) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
12 |
0.48 |
Binding ≤ 10μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B_MOUSE |
Q02152
|
Serotonin 2b (5-HT2b) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
12 |
0.48 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C_MOUSE |
P34968
|
Serotonin 2c (5-HT2c) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3A_MOUSE |
P23979
|
Serotonin 3a (5-HT3a) Receptor, Mouse |
32 |
0.46 |
Binding ≤ 10μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A_HUMAN |
P47898
|
Serotonin 5a (5-HT5a) Receptor, Human |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
39.8107171 |
0.45 |
Binding ≤ 10μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
6.3 |
0.50 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
8500 |
0.31 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
390 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
191 |
0.41 |
Functional ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
631 |
0.38 |
Functional ≤ 10μM
|
|
Z102275 |
Z102275
|
Hypothalamus |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.55 |
-7.08 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.14 |
8.88 |
-23.43 |
2 |
4 |
1 |
36 |
327.839 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.14 |
11.23 |
-89.99 |
3 |
4 |
2 |
38 |
328.847 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.02 |
-11.13 |
3 |
5 |
0 |
93 |
294.31 |
4 |
↓
|
|
|
|
Analogs
-
4982
-
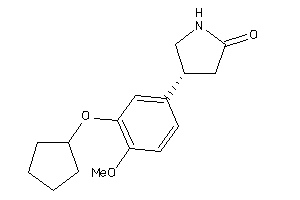
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.41 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
150 |
0.48 |
Binding ≤ 1μM
|
|
PDE1A_BOVIN |
P14100
|
Phosphodiesterase 1A, Bovin |
800 |
0.43 |
Binding ≤ 1μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
100 |
0.49 |
Binding ≤ 1μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
1.6 |
0.62 |
Binding ≤ 1μM
|
|
PDE4A_MOUSE |
O89084
|
Phosphodiesterase 4A, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
1.6 |
0.62 |
Binding ≤ 1μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
Q8VBU5_MOUSE |
Q8VBU5
|
Phosphodiesterase 4B, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
1.6 |
0.62 |
Binding ≤ 1μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
PDE4C_MOUSE |
Q3UEI1
|
Phosphodiesterase 4C, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
1.6 |
0.62 |
Binding ≤ 1μM
|
|
PDE4D_MOUSE |
Q01063
|
Phosphodiesterase 4D, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
720 |
0.43 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2 |
0.61 |
Binding ≤ 1μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
150 |
0.48 |
Binding ≤ 10μM
|
|
PDE1A_BOVIN |
P14100
|
Phosphodiesterase 1A, Bovin |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4A_MOUSE |
O89084
|
Phosphodiesterase 4A, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
100 |
0.49 |
Binding ≤ 10μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
Q8VBU5_MOUSE |
Q8VBU5
|
Phosphodiesterase 4B, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4C_MOUSE |
Q3UEI1
|
Phosphodiesterase 4C, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
1400 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D_MOUSE |
Q01063
|
Phosphodiesterase 4D, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
720 |
0.43 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
160 |
0.48 |
Functional ≤ 10μM
|
|
Z100489 |
Z100489
|
Eosinophils (Eosinophils) |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
225 |
0.47 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
160 |
0.48 |
Functional ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
1100 |
0.42 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
250 |
0.46 |
Functional ≤ 10μM
|
|
Z80566 |
Z80566
|
U-937 (Histiocytic Lymphoma Cells) |
1100 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.44 |
-11.23 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
2.11 |
-7.58 |
1 |
4 |
0 |
51 |
275.348 |
4 |
↓
|
|
|
|
Analogs
-
2000919
-
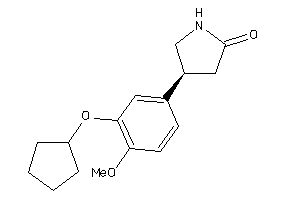
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A-1-E |
Phosphodiesterase 4A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.52 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C-1-E |
Phosphodiesterase 4C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3690 |
0.38 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
622 |
0.43 |
Binding ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.42 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
Q8VBU5-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B-1-E |
Phosphodiesterase 4B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D-1-E |
Phosphodiesterase 4D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 1), Other |
Other |
150 |
0.48 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
7 |
0.57 |
Binding ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z100489-1-O |
Eosinophils (Eosinophils) (cluster #1 Of 1), Other |
Other |
6500 |
0.36 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
340 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
660 |
0.43 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.52 |
Functional ≤ 10μM
|
|
Z80566-1-O |
U-937 (Histiocytic Lymphoma Cells) (cluster #1 Of 2), Other |
Other |
1230 |
0.41 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
150 |
0.48 |
Binding ≤ 1μM
|
|
PDE1A_BOVIN |
P14100
|
Phosphodiesterase 1A, Bovin |
800 |
0.43 |
Binding ≤ 1μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
100 |
0.49 |
Binding ≤ 1μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
162 |
0.48 |
Binding ≤ 1μM
|
|
PDE4A_MOUSE |
O89084
|
Phosphodiesterase 4A, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
231 |
0.46 |
Binding ≤ 1μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
Q8VBU5_MOUSE |
Q8VBU5
|
Phosphodiesterase 4B, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
1.6 |
0.62 |
Binding ≤ 1μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
PDE4C_MOUSE |
Q3UEI1
|
Phosphodiesterase 4C, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
622 |
0.43 |
Binding ≤ 1μM
|
|
PDE4D_MOUSE |
Q01063
|
Phosphodiesterase 4D, Mouse |
4 |
0.59 |
Binding ≤ 1μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
0.1 |
0.70 |
Binding ≤ 1μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
720 |
0.43 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2 |
0.61 |
Binding ≤ 1μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
150 |
0.48 |
Binding ≤ 10μM
|
|
PDE1A_BOVIN |
P14100
|
Phosphodiesterase 1A, Bovin |
800 |
0.43 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
1600 |
0.41 |
Binding ≤ 10μM
|
|
PDE4A_MOUSE |
O89084
|
Phosphodiesterase 4A, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
100 |
0.49 |
Binding ≤ 10μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
Q8VBU5_MOUSE |
Q8VBU5
|
Phosphodiesterase 4B, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
1600 |
0.41 |
Binding ≤ 10μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4C_MOUSE |
Q3UEI1
|
Phosphodiesterase 4C, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
1600 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
0.1 |
0.70 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
1600 |
0.41 |
Binding ≤ 10μM
|
|
PDE4D_MOUSE |
Q01063
|
Phosphodiesterase 4D, Mouse |
4 |
0.59 |
Binding ≤ 10μM
|
|
Q864F1_PIG |
Q864F1
|
Phosphodiesterase 5, Pig |
720 |
0.43 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
160 |
0.48 |
Functional ≤ 10μM
|
|
Z100489 |
Z100489
|
Eosinophils (Eosinophils) |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
225 |
0.47 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
4 |
0.59 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
160 |
0.48 |
Functional ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
625 |
0.43 |
Functional ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
1100 |
0.42 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
250 |
0.46 |
Functional ≤ 10μM
|
|
Z80566 |
Z80566
|
U-937 (Histiocytic Lymphoma Cells) |
1100 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.43 |
-11.26 |
1 |
4 |
0 |
48 |
275.348 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
MP2K1-3-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.42 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
5.85 |
-10.31 |
2 |
4 |
0 |
65 |
267.284 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A-1-E |
Melatonin Receptor 1A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
MTR1B-1-E |
Melatonin Receptor 1B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
MTR1C-1-E |
Melatonin Receptor 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1606 |
0.37 |
Binding ≤ 10μM
|
|
Z81346-1-O |
Melanophores (Melanophore Cells) (cluster #1 Of 1), Other |
Other |
2100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
8.44 |
-12.45 |
2 |
3 |
0 |
45 |
292.382 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.58 |
12.09 |
-8.52 |
0 |
4 |
0 |
42 |
382.891 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.58 |
12.29 |
-40.63 |
1 |
4 |
1 |
44 |
383.899 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
10000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
4.34 |
-11.23 |
4 |
5 |
0 |
91 |
256.096 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.04 |
4.52 |
-30.43 |
5 |
5 |
1 |
92 |
257.104 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
110 |
0.51 |
Binding ≤ 1μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
110 |
0.51 |
Binding ≤ 1μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
800 |
0.45 |
Binding ≤ 1μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
210 |
0.49 |
Binding ≤ 1μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
210 |
0.49 |
Binding ≤ 1μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
800 |
0.45 |
Binding ≤ 1μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
210 |
0.49 |
Binding ≤ 1μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
800 |
0.45 |
Binding ≤ 1μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
210 |
0.49 |
Binding ≤ 1μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
390 |
0.47 |
Binding ≤ 1μM
|
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
110 |
0.51 |
Binding ≤ 10μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
110 |
0.51 |
Binding ≤ 10μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
210 |
0.49 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
800 |
0.45 |
Binding ≤ 10μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
210 |
0.49 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
800 |
0.45 |
Binding ≤ 10μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
210 |
0.49 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
800 |
0.45 |
Binding ≤ 10μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
210 |
0.49 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
390 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
3.07 |
-12.5 |
1 |
5 |
0 |
64 |
268.219 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.40 |
1.22 |
-48.8 |
0 |
5 |
-1 |
67 |
267.211 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
610 |
0.40 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM |
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
365 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.46 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2870 |
0.35 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6740 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
958 |
0.38 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 24), Eukaryotic |
Eukaryotes |
69 |
0.46 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
88 |
0.45 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
10 |
0.51 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
14.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
118 |
0.44 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
17 |
0.49 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
15.7 |
0.50 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
39 |
0.47 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
39 |
0.47 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
173 |
0.43 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
181 |
0.43 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.2 |
0.57 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
0.3 |
0.61 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
2.1 |
0.55 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
22 |
0.49 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
26 |
0.48 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
26 |
0.48 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
26 |
0.48 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
26 |
0.48 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
958 |
0.38 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
610 |
0.40 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
1.9 |
0.55 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
12 |
0.50 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.51 |
Binding ≤ 1μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
365 |
0.41 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
14.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
14 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
2870 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
140 |
0.44 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
2870 |
0.35 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
118 |
0.44 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
15.7 |
0.50 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
17 |
0.49 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
39 |
0.47 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
39 |
0.47 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
173 |
0.43 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
181 |
0.43 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.2 |
0.57 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
0.3 |
0.61 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
22 |
0.49 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
2.1 |
0.55 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
26 |
0.48 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
26 |
0.48 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
26 |
0.48 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
26 |
0.48 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
958 |
0.38 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
610 |
0.40 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
2100 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
1.9 |
0.55 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
12 |
0.50 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.51 |
Binding ≤ 10μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
365 |
0.41 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
10 |
0.51 |
Functional ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
4 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6.89 |
-8.12 |
1 |
4 |
0 |
35 |
312.442 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
3996025
-
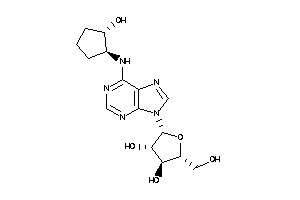
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.23 |
-13.5 |
-17.5 |
5 |
10 |
0 |
145 |
351.363 |
4 |
↓
|
|
|
|
Analogs
-
3784136
-
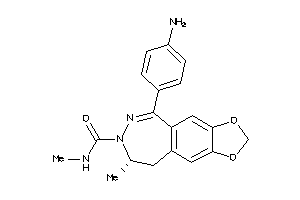
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
-2.64 |
-11.74 |
3 |
7 |
0 |
89 |
352.394 |
1 |
↓
|
|
|
|
Analogs
-
598361
-
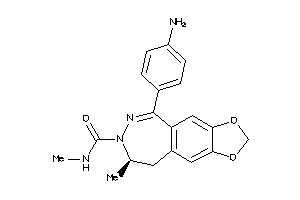
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
-2.67 |
-11.77 |
3 |
7 |
0 |
89 |
352.394 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.34 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
AVR2B-1-E |
Activin Receptor Type-2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.35 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.33 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.32 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
910 |
0.34 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.32 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.35 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2700 |
0.31 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.32 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.35 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.32 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.40 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.38 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.37 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.32 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
51 |
0.41 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.28 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.28 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.30 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.35 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.29 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.32 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.36 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.31 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.34 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.30 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Functional ≤ 10μM |
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
50 |
0.41 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.29 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B_HUMAN |
P36896
|
Activin Receptor Type-1B, Human |
950 |
0.34 |
Binding ≤ 1μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
120 |
0.39 |
Binding ≤ 1μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
42 |
0.41 |
Binding ≤ 1μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
530 |
0.35 |
Binding ≤ 1μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
59 |
0.40 |
Binding ≤ 1μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
170 |
0.38 |
Binding ≤ 1μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
510 |
0.35 |
Binding ≤ 1μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
210 |
0.37 |
Binding ≤ 1μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
13 |
0.44 |
Binding ≤ 1μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
100 |
0.39 |
Binding ≤ 1μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
80 |
0.40 |
Binding ≤ 1μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
80 |
0.40 |
Binding ≤ 1μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
170 |
0.38 |
Binding ≤ 1μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
270 |
0.37 |
Binding ≤ 1μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
28 |
0.42 |
Binding ≤ 1μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
790 |
0.34 |
Binding ≤ 1μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
620 |
0.35 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
53 |
0.41 |
Binding ≤ 1μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
640 |
0.35 |
Binding ≤ 1μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
150 |
0.38 |
Binding ≤ 1μM
|
|
ACV1B_HUMAN |
P36896
|
Activin Receptor Type-1B, Human |
950 |
0.34 |
Binding ≤ 10μM
|
|
AVR2B_HUMAN |
Q13705
|
Activin Receptor Type-2B, Human |
4800 |
0.30 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
2400 |
0.31 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
120 |
0.39 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
42 |
0.41 |
Binding ≤ 10μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
1700 |
0.32 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
530 |
0.35 |
Binding ≤ 10μM
|
|
KC1D_HUMAN |
P48730
|
Casein Kinase I Delta, Human |
59 |
0.40 |
Binding ≤ 10μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
170 |
0.38 |
Binding ≤ 10μM
|
|
KC1AL_HUMAN |
Q8N752
|
Casein Kinase I Isoform Alpha-like, Human |
1900 |
0.32 |
Binding ≤ 10μM
|
|
CTRO_HUMAN |
O14578
|
Citron Rho-interacting Kinase, Human |
510 |
0.35 |
Binding ≤ 10μM
|
|
DDR2_HUMAN |
Q16832
|
Discoidin Domain-containing Receptor 2, Human |
1600 |
0.32 |
Binding ≤ 10μM
|
|
MP2K2_HUMAN |
P36507
|
Dual Specificity Mitogen-activated Protein Kinase Kinase 2, Human |
9600 |
0.28 |
Binding ≤ 10μM
|
|
TTK_HUMAN |
P33981
|
Dual Specificity Protein Kinase TTK, Human |
4500 |
0.30 |
Binding ≤ 10μM
|
|
EPHA6_HUMAN |
Q9UF33
|
Ephrin Type-A Receptor 6, Human |
2200 |
0.32 |
Binding ≤ 10μM
|
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
1100 |
0.33 |
Binding ≤ 10μM
|
|
DDR1_HUMAN |
Q08345
|
Epithelial Discoidin Domain-containing Receptor 1, Human |
1100 |
0.33 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
2700 |
0.31 |
Binding ≤ 10μM
|
|
MK14_HUMAN |
Q16539
|
MAP Kinase P38 Alpha, Human |
13 |
0.44 |
Binding ≤ 10μM
|
|
MK11_HUMAN |
Q15759
|
MAP Kinase P38 Beta, Human |
100 |
0.39 |
Binding ≤ 10μM
|
|
MK13_HUMAN |
O15264
|
MAP Kinase P38 Delta, Human |
80 |
0.40 |
Binding ≤ 10μM
|
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
3300 |
0.31 |
Binding ≤ 10μM
|
|
KS6A1_HUMAN |
Q15418
|
Ribosomal Protein S6 Kinase Alpha 1, Human |
170 |
0.38 |
Binding ≤ 10μM
|
|
KS6A6_HUMAN |
Q9UK32
|
Ribosomal Protein S6 Kinase Alpha 6, Human |
270 |
0.37 |
Binding ≤ 10μM
|
|
NLK_HUMAN |
Q9UBE8
|
Serine/threonine Protein Kinase NLK, Human |
28 |
0.42 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
3200 |
0.31 |
Binding ≤ 10μM
|
|
ST32B_HUMAN |
Q9NY57
|
Serine/threonine-protein Kinase 32B, Human |
4900 |
0.30 |
Binding ≤ 10μM
|
|
STK36_HUMAN |
Q9NRP7
|
Serine/threonine-protein Kinase 36, Human |
3800 |
0.30 |
Binding ≤ 10μM
|
|
BRAF_HUMAN |
P15056
|
Serine/threonine-protein Kinase B-raf, Human |
4800 |
0.30 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
53 |
0.41 |
Binding ≤ 10μM
|
|
LATS2_HUMAN |
Q9NRM7
|
Serine/threonine-protein Kinase LATS2, Human |
1700 |
0.32 |
Binding ≤ 10μM
|
|
MRCKB_HUMAN |
Q9Y5S2
|
Serine/threonine-protein Kinase MRCK Beta, Human |
4200 |
0.30 |
Binding ≤ 10μM
|
|
MRCKG_HUMAN |
Q6DT37
|
Serine/threonine-protein Kinase MRCK Gamma, Human |
640 |
0.35 |
Binding ≤ 10μM
|
|
RAF1_HUMAN |
P04049
|
Serine/threonine-protein Kinase RAF, Human |
1900 |
0.32 |
Binding ≤ 10μM
|
|
RIPK2_HUMAN |
O43353
|
Serine/threonine-protein Kinase RIPK2, Human |
150 |
0.38 |
Binding ≤ 10μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
10000 |
0.28 |
Binding ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
4900 |
0.30 |
Binding ≤ 10μM
|
|
PTK6_HUMAN |
Q13882
|
Tyrosine-protein Kinase BRK, Human |
3000 |
0.31 |
Binding ≤ 10μM
|
|
FRK_HUMAN |
P42685
|
Tyrosine-protein Kinase FRK, Human |
3100 |
0.31 |
Binding ≤ 10μM
|
|
ADCK4_HUMAN |
Q96D53
|
Uncharacterized AarF Domain-containing Protein Kinase 4, Human |
4900 |
0.30 |
Binding ≤ 10μM
|
|
LOX5_HUMAN |
P09917
|
Arachidonate 5-lipoxygenase, Human |
50 |
0.41 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
50 |
0.41 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
6309.57344 |
0.29 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
1000 |
0.34 |
Functional ≤ 10μM
|
|
Z102116 |
Z102116
|
Toxoplasma Gondii RH |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.78 |
-41.1 |
2 |
4 |
0 |
62 |
331.35 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
9.26 |
-92.55 |
3 |
4 |
1 |
63 |
332.358 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.78 |
7.25 |
-9.85 |
2 |
4 |
0 |
62 |
331.35 |
3 |
↓
|
|
|
|
Analogs
-
839816
-
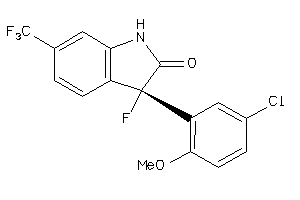
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.88 |
-8.59 |
1 |
3 |
0 |
38 |
359.706 |
3 |
↓
|
|
|
|
Analogs
-
538646
-
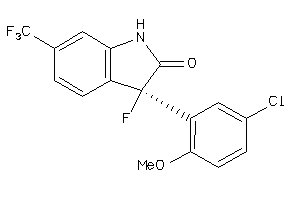
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.86 |
-8.57 |
1 |
3 |
0 |
38 |
359.706 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
597419
-
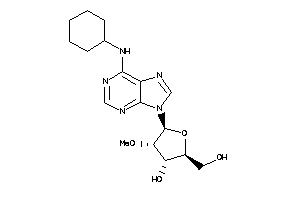
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
76 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
-8.58 |
-13.38 |
3 |
9 |
0 |
114 |
363.418 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
2.1 |
-14.41 |
0 |
8 |
0 |
80 |
384.436 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
287 |
0.35 |
Binding ≤ 1μM
|
|
AA1R_BOVIN |
P28190
|
Adenosine A1 Receptor, Bovin |
357 |
0.35 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
121 |
0.37 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
2.3 |
0.47 |
Binding ≤ 1μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
AA1R_BOVIN |
P28190
|
Adenosine A1 Receptor, Bovin |
357 |
0.35 |
Binding ≤ 10μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
121 |
0.37 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
287 |
0.35 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
1900 |
0.31 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
12 |
0.43 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
12 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
10.81 |
-11.79 |
2 |
8 |
0 |
100 |
345.366 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
830 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
11.61 |
-28.27 |
0 |
3 |
0 |
43 |
412.439 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
9.46 |
-8.08 |
1 |
5 |
0 |
45 |
395.547 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
437 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1393 |
0.28 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1393 |
0.28 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1393 |
0.28 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
897 |
0.29 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
176 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
9 |
-8.74 |
1 |
5 |
0 |
45 |
395.547 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
3.46 |
-10.52 |
3 |
6 |
0 |
86 |
311.341 |
3 |
↓
|
|
|
|
Analogs
-
512
-
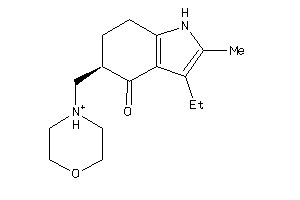
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
5.69 |
-11.34 |
1 |
4 |
0 |
45 |
276.38 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
7.04 |
-48.98 |
1 |
4 |
1 |
43 |
277.388 |
3 |
↓
|
|
|
|
Analogs
-
512
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
265 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
5.69 |
-11.35 |
1 |
4 |
0 |
45 |
276.38 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
6.82 |
-50.27 |
1 |
4 |
1 |
43 |
277.388 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.50 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.42 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
55 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
64 |
0.44 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
64 |
0.44 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
11 |
0.48 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
12.5892541 |
0.48 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
758 |
0.37 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
120 |
0.42 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
758 |
0.37 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
110 |
0.42 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
758 |
0.37 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
20.8929613 |
0.47 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
6 |
0.50 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 1μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
22 |
0.47 |
Binding ≤ 1μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
22 |
0.47 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
64 |
0.44 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
11 |
0.48 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
12.5892541 |
0.48 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
120 |
0.42 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
110 |
0.42 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
758 |
0.37 |
Binding ≤ 10μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
20.8929613 |
0.47 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
6 |
0.50 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.77827941 |
0.53 |
Binding ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
22 |
0.47 |
Binding ≤ 10μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
22 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.56 |
-6.67 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
1 |
-13.69 |
2 |
6 |
0 |
90 |
280.334 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
1.48 |
-36.6 |
1 |
6 |
-1 |
88 |
279.326 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
10.79 |
-11.94 |
2 |
3 |
0 |
45 |
334.463 |
8 |
↓
|
|