|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
13.94 |
-59.04 |
3 |
5 |
1 |
78 |
518.077 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.13 |
4.89 |
-11.19 |
1 |
6 |
0 |
84 |
219.628 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.13 |
4.3 |
-12.26 |
1 |
6 |
0 |
84 |
219.628 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
10 |
-42.36 |
2 |
2 |
1 |
29 |
311.836 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PARP1-1-E |
Poly [ADP-ribose] Polymerase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
7.89 |
-53.46 |
2 |
5 |
1 |
51 |
321.404 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.42 |
8.32 |
-91.18 |
3 |
5 |
2 |
53 |
322.412 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
2 |
0.39 |
Binding ≤ 1μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
3 |
0.38 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
3 |
0.38 |
Binding ≤ 1μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
600 |
0.28 |
Binding ≤ 1μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
4.2 |
0.38 |
Binding ≤ 1μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
240 |
0.30 |
Binding ≤ 1μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
39 |
0.33 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
390 |
0.29 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
2 |
0.39 |
Binding ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
3 |
0.38 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
3 |
0.38 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
1050 |
0.27 |
Binding ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
600 |
0.28 |
Binding ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
4.2 |
0.38 |
Binding ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
240 |
0.30 |
Binding ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
39 |
0.33 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
390 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
-0.73 |
-49.96 |
4 |
7 |
1 |
92 |
422.505 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
-0.17 |
-87.78 |
3 |
7 |
0 |
98 |
421.497 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.83 |
-0.17 |
-87.78 |
3 |
7 |
0 |
94 |
421.497 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
7.67 |
-48.85 |
2 |
9 |
1 |
110 |
498.647 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.69 |
6.65 |
-22.44 |
1 |
9 |
0 |
109 |
497.639 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
7.61 |
-53.6 |
3 |
5 |
1 |
66 |
333.415 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.65 |
6.88 |
-92.85 |
7 |
7 |
2 |
113 |
438.963 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.51 |
6.24 |
-14.79 |
2 |
5 |
0 |
61 |
332.407 |
3 |
↓
|
|
|
|
Analogs
-
43190979
-
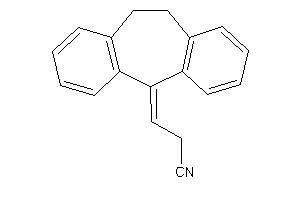
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5B-1-E |
Serotonin 5b (5-HT5b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADCY1-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.48 |
Binding ≤ 10μM
|
|
ADCY2-2-E |
Adenylate Cyclase Type II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.48 |
Binding ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.48 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.49 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
89 |
0.47 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
206 |
0.45 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.45 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8500 |
0.34 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.41 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Functional ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
196 |
0.45 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
300 |
0.43 |
Binding ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
5400 |
0.35 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
61 |
0.48 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADCY2_HUMAN |
Q08462
|
Adenylate Cyclase Type II, Human |
53 |
0.49 |
Binding ≤ 1μM
|
|
ADCY8_HUMAN |
P40145
|
Adenylate Cyclase Type VIII, Human |
53 |
0.49 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.1 |
0.67 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADRB2_RAT |
P10608
|
Beta-2 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 1μM
|
|
ADCY1_HUMAN |
Q08828
|
Brain Adenylate Cyclase 1, Human |
53 |
0.49 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
89 |
0.47 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
196 |
0.45 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
206 |
0.45 |
Binding ≤ 1μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
170 |
0.45 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.12 |
0.60 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
0.1 |
0.67 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
300 |
0.43 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT4R_RAT |
Q62758
|
Serotonin 4 (5-HT4) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT5A_RAT |
P35364
|
Serotonin 5a (5-HT5a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT5B_RAT |
P35365
|
Serotonin 5b (5-HT5b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT6R_RAT |
P31388
|
Serotonin 6 (5-HT6) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
44 |
0.49 |
Binding ≤ 1μM
|
|
ADCY2_HUMAN |
Q08462
|
Adenylate Cyclase Type II, Human |
53 |
0.49 |
Binding ≤ 10μM
|
|
ADCY8_HUMAN |
P40145
|
Adenylate Cyclase Type VIII, Human |
53 |
0.49 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
0.1 |
0.67 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADRB2_RAT |
P10608
|
Beta-2 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
41 |
0.49 |
Binding ≤ 10μM
|
|
ADCY1_HUMAN |
Q08828
|
Brain Adenylate Cyclase 1, Human |
53 |
0.49 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
89 |
0.47 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
196 |
0.45 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
206 |
0.45 |
Binding ≤ 10μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
170 |
0.45 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10000 |
0.33 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.12 |
0.60 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
0.1 |
0.67 |
Binding ≤ 10μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
300 |
0.43 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
3000 |
0.37 |
Binding ≤ 10μM
|
|
ACM5_RAT |
P08911
|
Muscarinic Acetylcholine Receptor M5, Rat |
3000 |
0.37 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
2500 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT4R_RAT |
Q62758
|
Serotonin 4 (5-HT4) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5A_RAT |
P35364
|
Serotonin 5a (5-HT5a) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5B_RAT |
P35365
|
Serotonin 5b (5-HT5b) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT6R_RAT |
P31388
|
Serotonin 6 (5-HT6) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
44 |
0.49 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
8500 |
0.34 |
Binding ≤ 10μM
|
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
10000 |
0.33 |
Binding ≤ 10μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
10000 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
10000 |
0.33 |
Functional ≤ 10μM
|
|
HRH2_HUMAN |
P25021
|
Histamine H2 Receptor, Human |
660 |
0.41 |
Functional ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
2900 |
0.37 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
12.75 |
-36.66 |
1 |
1 |
1 |
4 |
278.419 |
3 |
↓
|
|
|
|
Analogs
-
3794622
-
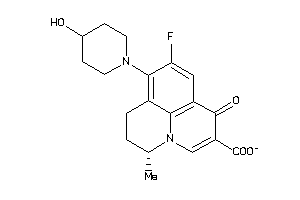
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.49 |
8.73 |
-70.79 |
1 |
6 |
-1 |
86 |
359.377 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.50 |
6.08 |
-32.34 |
2 |
6 |
0 |
89 |
360.385 |
1 |
↓
|
|
|
|
Analogs
-
603195
-
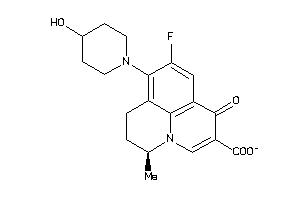
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.49 |
8.73 |
-70.73 |
1 |
6 |
-1 |
86 |
359.377 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.50 |
6.06 |
-33.01 |
2 |
6 |
0 |
89 |
360.385 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
5.42 |
-40.52 |
3 |
3 |
1 |
40 |
231.319 |
1 |
↓
|
|
|
|
Analogs
-
32108748
-
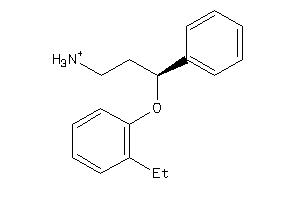
-
39298020
-
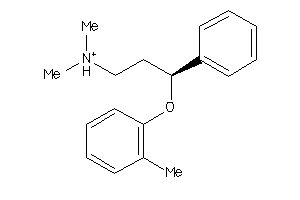
-
39298021
-
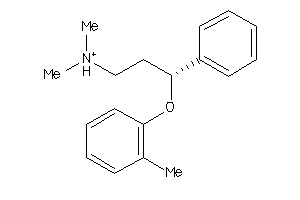
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2100 |
0.42 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4383 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4186 |
0.40 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.61 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3100 |
0.41 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
77 |
0.52 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1585 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
4 |
0.62 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
2.03 |
0.64 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
0.7 |
0.67 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
43 |
0.54 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
180 |
0.50 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
4 |
0.62 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
1400 |
0.43 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
1080 |
0.44 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
2100 |
0.42 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
4383 |
0.39 |
Binding ≤ 10μM
|
|
OPRK_CAVPO |
P41144
|
Kappa Opioid Receptor, Guinea Pig |
4383 |
0.39 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
4186 |
0.40 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
4186 |
0.40 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
0.7 |
0.67 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
2.03 |
0.64 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
1500 |
0.43 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
180 |
0.50 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
4 |
0.62 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1584.89319 |
0.43 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
25.1 |
0.56 |
Functional ≤ 10μM
|
|
CP2D6_HUMAN |
P10635
|
Cytochrome P450 2D6, Human |
2000 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.89 |
-40.02 |
2 |
2 |
1 |
26 |
256.369 |
6 |
↓
|
|
|
|
Analogs
-
6130066
-
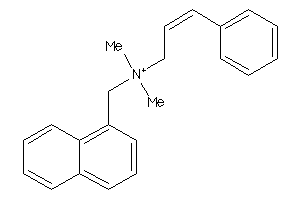
-
7997897
-
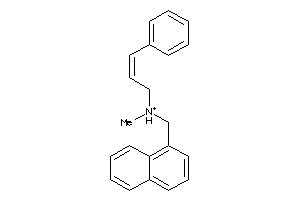
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
880 |
0.39 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
310 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
13.58 |
-41.74 |
1 |
1 |
1 |
4 |
288.414 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
0.33 |
-14.7 |
2 |
4 |
0 |
59 |
113.12 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.74 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Functional ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
12 |
0.79 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
1.6 |
0.88 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
21 |
0.77 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
0.87 |
0.91 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
0.78 |
0.91 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
130 |
0.69 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
1.6 |
0.88 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
1900 |
0.57 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
12 |
0.79 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
0.78 |
0.91 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
0.87 |
0.91 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
130 |
0.69 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
5 |
0.83 |
Functional ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
12 |
0.79 |
Functional ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
1.5 |
0.88 |
Functional ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
15 |
0.78 |
Functional ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
4700 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
3.81 |
-44.37 |
4 |
3 |
1 |
56 |
212.342 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.09 |
4.24 |
-88 |
5 |
3 |
2 |
57 |
213.35 |
3 |
↓
|
|
|
|
Analogs
-
4831675
-
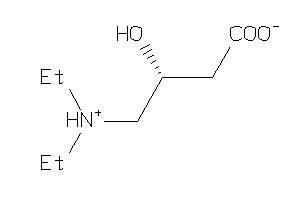
-
4831676
-
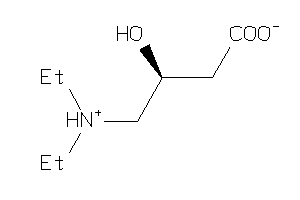
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.57 |
4.42 |
-54.16 |
1 |
4 |
0 |
60 |
161.201 |
4 |
↓
|
|
|
|
Analogs
-
518912
-
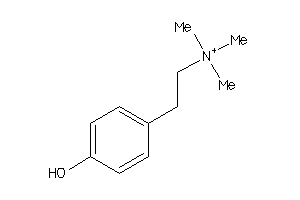
-
34363810
-
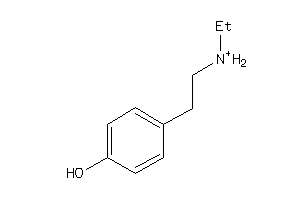
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
4.38 |
-38.98 |
2 |
2 |
1 |
25 |
166.244 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.67 |
1.91 |
-3.67 |
1 |
2 |
0 |
23 |
165.236 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.04 |
5.92 |
-18.76 |
5 |
10 |
0 |
153 |
504.501 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3881666
-
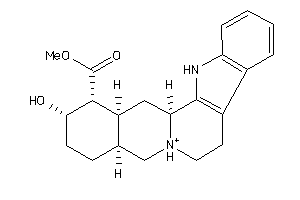
-
3953891
-
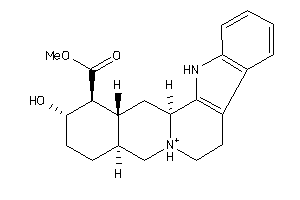
-
3977785
-
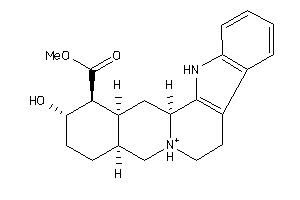
-
4398249
-
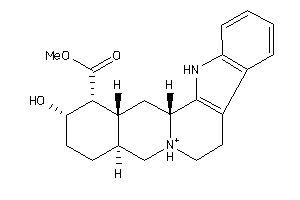
-
4535881
-
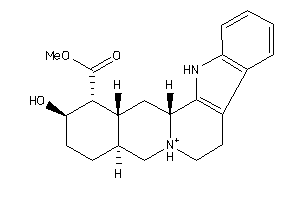
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1622 |
0.31 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
95 |
0.38 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1057 |
0.32 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
966 |
0.32 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
289 |
0.35 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
49 |
0.39 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
54 |
0.39 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
54 |
0.39 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.40 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2430 |
0.30 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
165 |
0.37 |
Functional ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
165 |
0.37 |
Functional ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
165 |
0.37 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.44 |
ADME/T ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.38 |
ADME/T ≤ 10μM
|
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
360 |
0.35 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
1100 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
505 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
95 |
0.38 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1000 |
0.32 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
200 |
0.36 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
1.1 |
0.48 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
1.6 |
0.47 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
52 |
0.39 |
Binding ≤ 1μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
49 |
0.39 |
Binding ≤ 1μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA2A_PIG |
P18871
|
Alpha-2a Adrenergic Receptor, Pig |
4.4 |
0.45 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 1μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA2B_BOVIN |
O77700
|
Alpha-2b Adrenergic Receptor, Bovin |
49 |
0.39 |
Binding ≤ 1μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 1μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
45 |
0.40 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
95 |
0.38 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1000 |
0.32 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
1057 |
0.32 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
200 |
0.36 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
1.1 |
0.48 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
52 |
0.39 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
1.6 |
0.47 |
Binding ≤ 10μM
|
|
ADA2A_MOUSE |
Q01338
|
Alpha-2a Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2A_PIG |
P18871
|
Alpha-2a Adrenergic Receptor, Pig |
4.4 |
0.45 |
Binding ≤ 10μM
|
|
ADA2A_BOVIN |
Q28838
|
Alpha-2a Adrenergic Receptor, Bovin |
49 |
0.39 |
Binding ≤ 10μM
|
|
ADA2B_BOVIN |
O77700
|
Alpha-2b Adrenergic Receptor, Bovin |
49 |
0.39 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2B_MOUSE |
P30545
|
Alpha-2b Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2C_MOUSE |
Q01337
|
Alpha-2c Adrenergic Receptor, Mouse |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
0.6 |
0.50 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
0.4 |
0.51 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
2000 |
0.31 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1125 |
0.32 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
2000 |
0.31 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
2000 |
0.31 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
2000 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1621.8101 |
0.31 |
Binding ≤ 10μM
|
|
5HT2C_MOUSE |
P34968
|
Serotonin 2c (5-HT2c) Receptor, Mouse |
10000 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C_RAT |
P22002
|
Voltage-gated L-type Calcium Channel Alpha-1C Subunit, Rat |
45 |
0.40 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
165 |
0.37 |
Functional ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
165 |
0.37 |
Functional ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
165 |
0.37 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
1100 |
0.32 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1995.26231 |
0.31 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
505 |
0.34 |
Functional ≤ 10μM
|
|
CP2D6_HUMAN |
P10635
|
Cytochrome P450 2D6, Human |
180 |
0.36 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
8.17 |
-43.2 |
3 |
5 |
1 |
67 |
355.458 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.22 |
6.05 |
-10.14 |
2 |
5 |
0 |
66 |
354.45 |
2 |
↓
|
|
|
|
Analogs
-
3651083
-
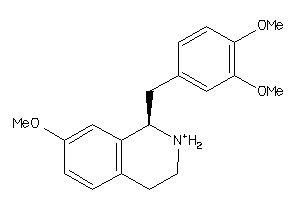
-
3651084
-
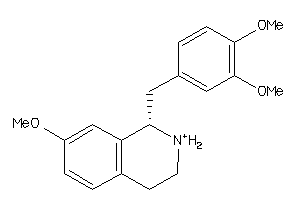
-
4262569
-
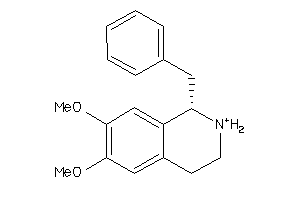
-
4262570
-
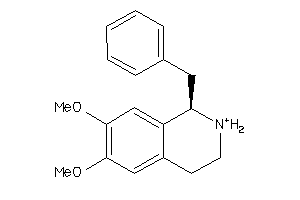
-
5811656
-
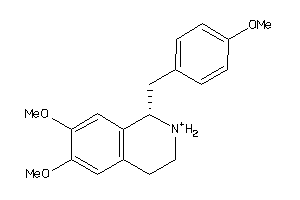
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.38 |
-51.89 |
2 |
5 |
1 |
54 |
344.431 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
6.21 |
-11.34 |
1 |
5 |
0 |
49 |
343.423 |
6 |
↓
|
|
|
|
Analogs
-
3651083
-

-
3651084
-

-
4262569
-

-
4262570
-

-
5811656
-

Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
1000 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.39 |
-51.14 |
2 |
5 |
1 |
54 |
344.431 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
6.11 |
-10.65 |
1 |
5 |
0 |
49 |
343.423 |
6 |
↓
|
|
|
|
Analogs
-
403588
-
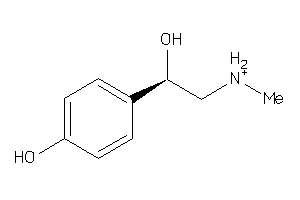
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
-0.78 |
-43.1 |
4 |
3 |
1 |
57 |
168.216 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
-2.29 |
-5.42 |
3 |
3 |
0 |
52 |
167.208 |
3 |
↓
|
|
|
|
Analogs
-
733
-
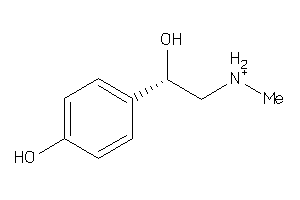
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
-0.72 |
-42.44 |
4 |
3 |
1 |
57 |
168.216 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.43 |
-2.17 |
-5.01 |
3 |
3 |
0 |
52 |
167.208 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
5273809
-
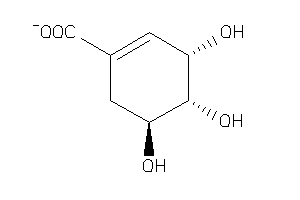
-
8681449
-
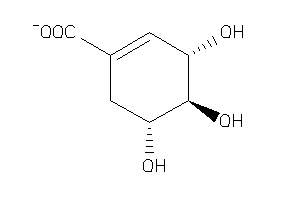
-
9214237
-
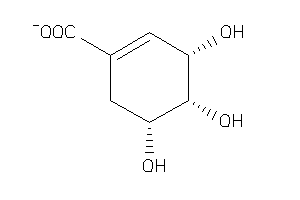
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.57 |
-7.38 |
-48.13 |
3 |
5 |
-1 |
100 |
173.144 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.80 |
-4.36 |
-57.03 |
3 |
4 |
0 |
85 |
125.149 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
10 |
0.66 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
80 |
0.58 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
9900 |
0.41 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
700 |
0.51 |
Functional ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
5000 |
0.44 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
390 |
0.53 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
9.7 |
0.66 |
Binding ≤ 1μM
|
|
DRTS_PLAFK |
P13922
|
Dihydrofolate Reductase, Plafk |
0.19 |
0.80 |
Binding ≤ 1μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
1.2 |
0.73 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
2800 |
0.46 |
Binding ≤ 10μM
|
|
DRTS_PLAFK |
P13922
|
Dihydrofolate Reductase, Plafk |
0.19 |
0.80 |
Binding ≤ 10μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
1.2 |
0.73 |
Binding ≤ 10μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1500 |
0.48 |
Binding ≤ 10μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
390 |
0.53 |
Binding ≤ 10μM
|
|
DRTS_PLAFK |
P13922
|
Dihydrofolate Reductase, Plafk |
80 |
0.58 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1.2 |
0.73 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
9900 |
0.41 |
Functional ≤ 10μM |
|
Z50339 |
Z50339
|
Pneumocystis Carinii |
0.1 |
0.82 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
0.1 |
0.82 |
Functional ≤ 10μM
|
|
Z50418 |
Z50418
|
Trypanosoma Brucei |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
5000 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
6.53 |
-28.58 |
5 |
4 |
1 |
79 |
249.725 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
6.08 |
-5.33 |
4 |
4 |
0 |
78 |
248.717 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.30 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
8 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
16.04 |
-72.01 |
2 |
6 |
1 |
64 |
514.685 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
10.77 |
-65.17 |
1 |
6 |
-1 |
92 |
446.882 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.93 |
11.38 |
-50.45 |
3 |
7 |
1 |
74 |
352.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC3-3-E |
Histone Deacetylase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9000 |
1.18 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
3.61 |
-43.26 |
0 |
2 |
-1 |
40 |
87.098 |
2 |
↓
|
|
|
|
Analogs
-
34676244
-
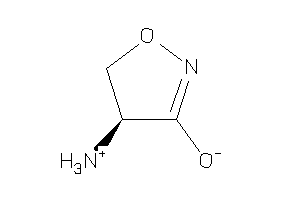
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7370 |
1.03 |
Binding ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
2300 |
1.13 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-5.16 |
-42.02 |
3 |
4 |
0 |
72 |
102.093 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-1.71 |
-6.26 |
-6.38 |
3 |
4 |
0 |
68 |
102.093 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.71 |
-5.45 |
-42.88 |
2 |
4 |
-1 |
71 |
101.085 |
0 |
↓
|
|
|
|
Analogs
-
525860
-
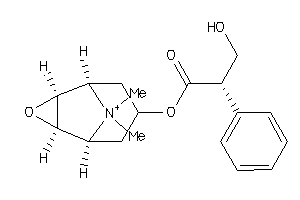
-
1088597
-
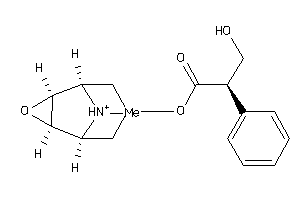
-
2034193
-
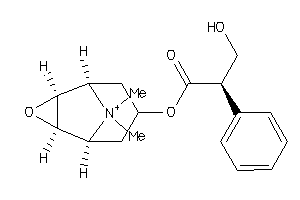
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.323593657 |
0.58 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
0.177827941 |
0.59 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
0.134896288 |
0.60 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
0.141253754 |
0.60 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
0.208929613 |
0.59 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.323593657 |
0.58 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
0.177827941 |
0.59 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
0.134896288 |
0.60 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
0.141253754 |
0.60 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
0.208929613 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.93 |
7.41 |
-35.57 |
1 |
5 |
1 |
59 |
318.393 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
7.59 |
-35 |
3 |
7 |
1 |
80 |
395.483 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.72 |
7.1 |
-11.67 |
2 |
7 |
0 |
79 |
394.475 |
5 |
↓
|
|
|
|
Analogs
-
1530690
-
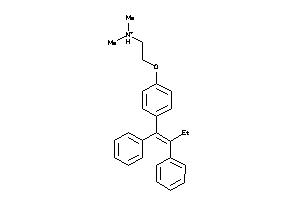
-
1849472
-
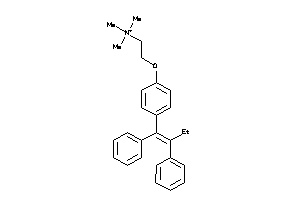
-
8602413
-
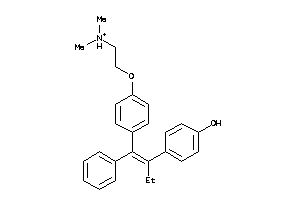
-
13319948
-
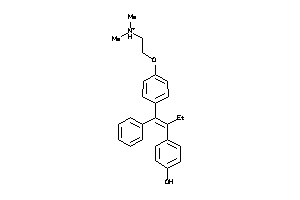
-
44699472
-
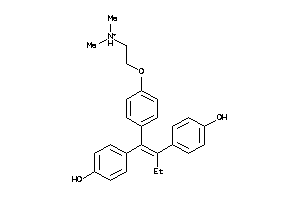
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1585 |
0.29 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.26 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.26 |
Binding ≤ 10μM
|
|
PDE1B-1-E |
Phosphodiesterase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6750 |
0.26 |
Binding ≤ 10μM
|
|
PDE1C-1-E |
Phosphodiesterase 1C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.26 |
Binding ≤ 10μM
|
|
Q8AWE3-1-E |
Serine-threonine Protein Phosphatase 2A Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.37 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
904 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
904 |
0.30 |
Functional ≤ 10μM
|
|
CP2B6-2-E |
Cytochrome P450 2B6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.30 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.33 |
ADME/T ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
1500 |
0.29 |
Binding ≤ 10μM
|
|
Z100267-1-O |
Anti-estrogen Binding Site (AEBS) (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.26 |
Functional ≤ 10μM
|
|
Z80132-2-O |
EL4 (Thymoma Cells) (cluster #2 Of 2), Other |
Other |
535 |
0.31 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
904 |
0.30 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z80231-2-O |
MCF7S (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
7200 |
0.26 |
Functional ≤ 10μM
|
|
Z80296-1-O |
MVLN (cluster #1 Of 1), Other |
Other |
300 |
0.33 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
1000 |
0.30 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
420 |
0.32 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
9860 |
0.25 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
10000 |
0.25 |
ADME/T ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EBP_HUMAN |
Q15125
|
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase, Human |
5 |
0.42 |
Binding ≤ 1μM
|
|
Z100267 |
Z100267
|
Anti-estrogen Binding Site (AEBS) |
1 |
0.45 |
Binding ≤ 1μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
100 |
0.35 |
Binding ≤ 1μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
11.9 |
0.40 |
Binding ≤ 1μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
100 |
0.35 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
35 |
0.37 |
Binding ≤ 1μM
|
|
EBP_HUMAN |
Q15125
|
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase, Human |
5 |
0.42 |
Binding ≤ 10μM
|
|
Z100267 |
Z100267
|
Anti-estrogen Binding Site (AEBS) |
1 |
0.45 |
Binding ≤ 10μM
|
|
ERG2_YEAST |
P32352
|
C-8 Sterol Isomerase, Yeast |
1500 |
0.29 |
Binding ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
100 |
0.35 |
Binding ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
11.9 |
0.40 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
1584.89319 |
0.29 |
Binding ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
100 |
0.35 |
Binding ≤ 10μM
|
|
PDE1A_HUMAN |
P54750
|
Phosphodiesterase 1A, Human |
6750 |
0.26 |
Binding ≤ 10μM
|
|
PDE1B_RAT |
Q01066
|
Phosphodiesterase 1B, Rat |
6750 |
0.26 |
Binding ≤ 10μM
|
|
PDE1B_HUMAN |
Q01064
|
Phosphodiesterase 1B, Human |
6750 |
0.26 |
Binding ≤ 10μM
|
|
PDE1C_HUMAN |
Q14123
|
Phosphodiesterase 1C, Human |
6750 |
0.26 |
Binding ≤ 10μM
|
|
O77823_PIG |
O77823
|
Phosphodiesterase 4A, Pig |
6800 |
0.26 |
Binding ≤ 10μM
|
|
Q8AWE3_CHICK |
Q8AWE3
|
Serine-threonine Protein Phosphatase 2A Regulatory Subunit, Chick |
3400 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
35 |
0.37 |
Binding ≤ 10μM
|
|
Z80132 |
Z80132
|
EL4 (Thymoma Cells) |
535 |
0.31 |
Functional ≤ 10μM
|
|
ESR1_HUMAN |
P03372
|
Estrogen Receptor Alpha, Human |
2540 |
0.28 |
Functional ≤ 10μM
|
|
ESR2_HUMAN |
Q92731
|
Estrogen Receptor Beta, Human |
1660 |
0.29 |
Functional ≤ 10μM
|
|
Z81068 |
Z81068
|
Ishikawa (Uterine Carcinoma Cells) |
170 |
0.34 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
0.11 |
0.50 |
Functional ≤ 10μM
|
|
Z80227 |
Z80227
|
MCF7-ADR (Breast Carcinoma Cells) |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z80231 |
Z80231
|
MCF7S (Breast Carcinoma Cells) |
7200 |
0.26 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
10000 |
0.25 |
Functional ≤ 10μM
|
|
Z81245 |
Z81245
|
MDA-MB-435 (Breast Carcinoma Cells) |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z80296 |
Z80296
|
MVLN |
300 |
0.33 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.25 |
Functional ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
1000 |
0.30 |
Functional ≤ 10μM
|
|
CP2B6_HUMAN |
P20813
|
Cytochrome P450 2B6, Human |
900 |
0.30 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
200 |
0.33 |
ADME/T ≤ 10μM
|
|
Z80936 |
Z80936
|
HEK293 (Embryonic Kidney Fibroblasts) |
10000 |
0.25 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.06 |
15.81 |
-35.9 |
1 |
2 |
1 |
14 |
372.532 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.06 |
13.36 |
-4.97 |
0 |
2 |
0 |
12 |
371.524 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.52 |
3.37 |
-9.16 |
1 |
7 |
0 |
97 |
211.177 |
5 |
↓
|
|
|
|
Analogs
-
643055
-
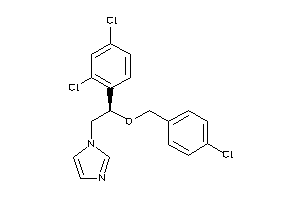
-
896740
-
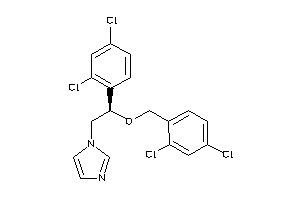
-
3872945
-
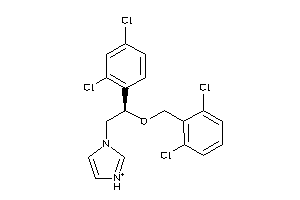
-
3944782
-
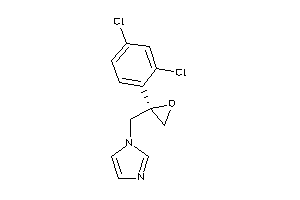
-
596881
-
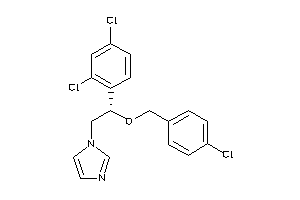
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
5 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
GRM6-2-E |
Metabotropic Glutamate Receptor 6 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
851 |
0.34 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
2 |
0.49 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.37 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
330 |
0.36 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 1μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 1μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 10μM
|
|
Z50416 |
Z50416
|
Aspergillus Fumigatus |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442 |
Z50442
|
Candida Albicans |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50443 |
Z50443
|
Candida Glabrata |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50408 |
Z50408
|
Issatchenkia Orientalis |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409 |
Z50409
|
Kluyveromyces Marxianus |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50459 |
Z50459
|
Leishmania Donovani |
6000 |
0.29 |
Functional ≤ 10μM
|
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
7800 |
0.29 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
2.03 |
0.49 |
Functional ≤ 10μM
|
|
Z102121 |
Z102121
|
Trichophyton Mentagrophytes |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50046 |
Z50046
|
Trichophyton Quinckeanum |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50452 |
Z50452
|
Trichophyton Rubrum |
140 |
0.38 |
Functional ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
180 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.15 |
-35.65 |
1 |
3 |
1 |
28 |
417.143 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
13.63 |
-6.4 |
0 |
3 |
0 |
27 |
416.135 |
6 |
↓
|
|
|
|
Analogs
-
3944782
-

-
596881
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
5 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
|
GRM6-2-E |
Metabotropic Glutamate Receptor 6 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
851 |
0.34 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
2 |
0.49 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.37 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
330 |
0.36 |
Functional ≤ 10μM
|
|
Z50459-2-O |
Leishmania Donovani (cluster #2 Of 8), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 1μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 1μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 1μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 1μM
|
|
AMPC_ECOLI |
P00811
|
Beta-lactamase, Ecoli |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
243 |
0.37 |
Binding ≤ 10μM
|
|
CP19A_RAT |
P22443
|
Cytochrome P450 19A1, Rat |
400 |
0.36 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
600 |
0.35 |
Binding ≤ 10μM
|
|
CP51A_HUMAN |
Q16850
|
Cytochrome P450 51, Human |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP51_MYCTU |
P0A512
|
Lanosterol 14-alpha Demethylase, Myctu |
200 |
0.38 |
Binding ≤ 10μM
|
|
Z50416 |
Z50416
|
Aspergillus Fumigatus |
1900 |
0.32 |
Functional ≤ 10μM
|
|
Z50442 |
Z50442
|
Candida Albicans |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50443 |
Z50443
|
Candida Glabrata |
120 |
0.39 |
Functional ≤ 10μM
|
|
Z50408 |
Z50408
|
Issatchenkia Orientalis |
1400 |
0.33 |
Functional ≤ 10μM
|
|
Z50409 |
Z50409
|
Kluyveromyces Marxianus |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z50459 |
Z50459
|
Leishmania Donovani |
6000 |
0.29 |
Functional ≤ 10μM
|
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
6500 |
0.29 |
Functional ≤ 10μM
|
|
MDR1_MOUSE |
P06795
|
P-glycoprotein 1, Mouse |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR1_HUMAN |
P08183
|
P-glycoprotein 1, Human |
2000 |
0.32 |
Functional ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
7800 |
0.29 |
Functional ≤ 10μM
|
|
Z50038 |
Z50038
|
Plasmodium Yoelii Yoelii |
2.03 |
0.49 |
Functional ≤ 10μM
|
|
Z102121 |
Z102121
|
Trichophyton Mentagrophytes |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50046 |
Z50046
|
Trichophyton Quinckeanum |
790 |
0.34 |
Functional ≤ 10μM
|
|
Z50452 |
Z50452
|
Trichophyton Rubrum |
140 |
0.38 |
Functional ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
6000 |
0.29 |
ADME/T ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
180 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
13.6 |
-35.01 |
1 |
3 |
1 |
28 |
417.143 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
13.09 |
-6.46 |
0 |
3 |
0 |
27 |
416.135 |
6 |
↓
|
|